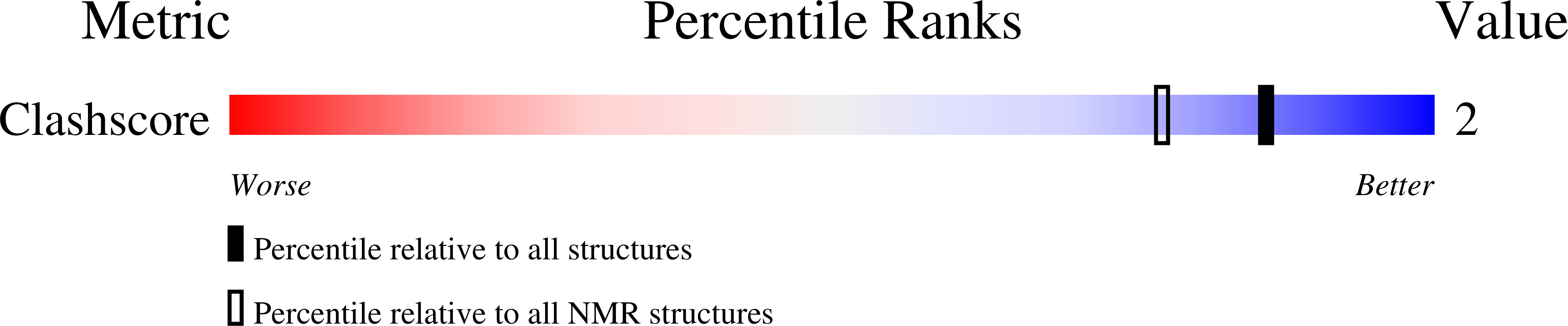
Deposition Date
2001-08-29
Release Date
2002-05-22
Last Version Date
2024-05-22
Entry Detail
Method Details:
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
10
Selection Criteria:
Lowest target function (a weighted sum of conformational energy and restraint energy).