Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
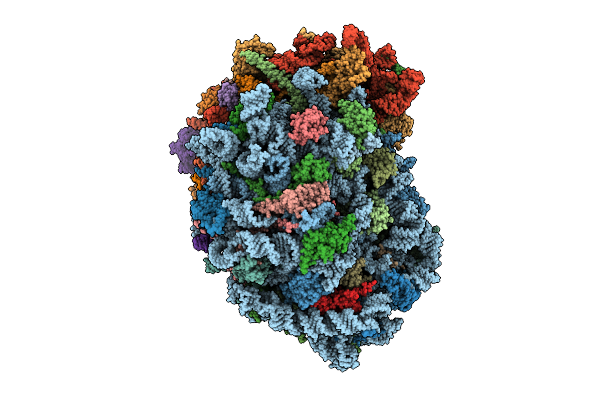 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
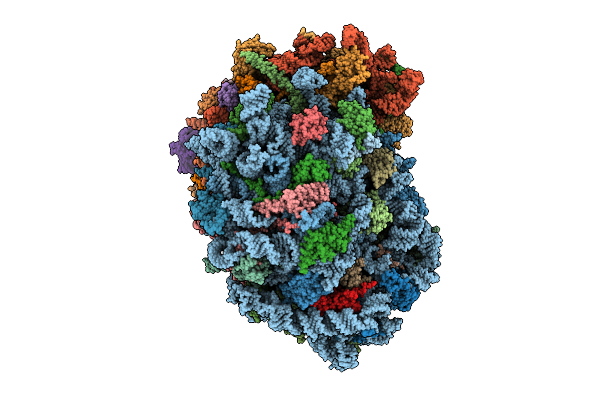 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
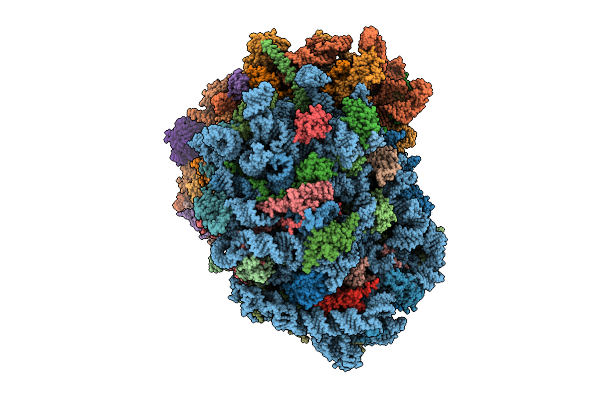 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, ZN, SPM, SPD |
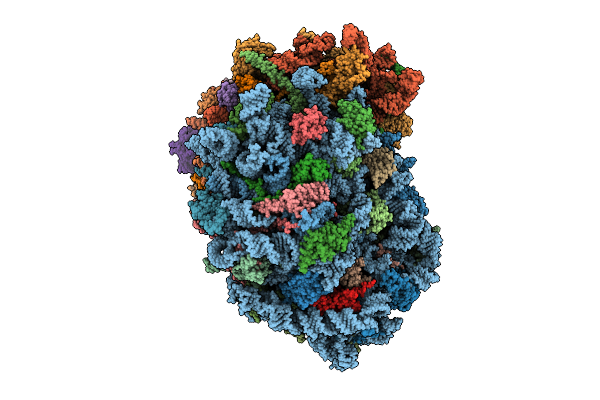 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
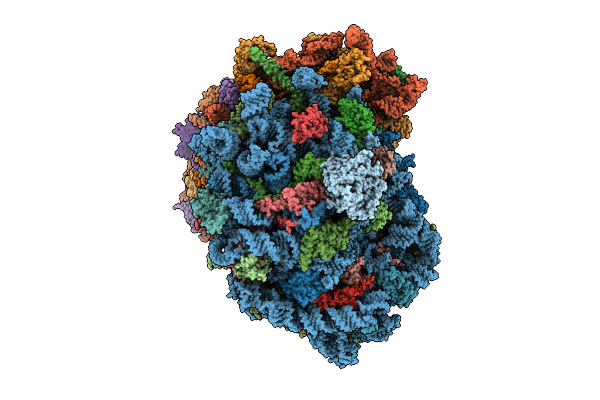 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
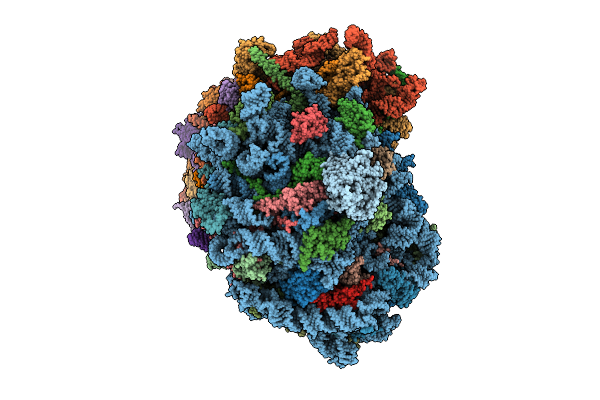 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
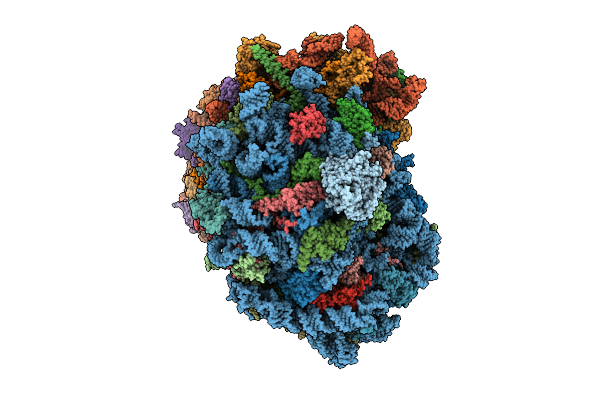 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
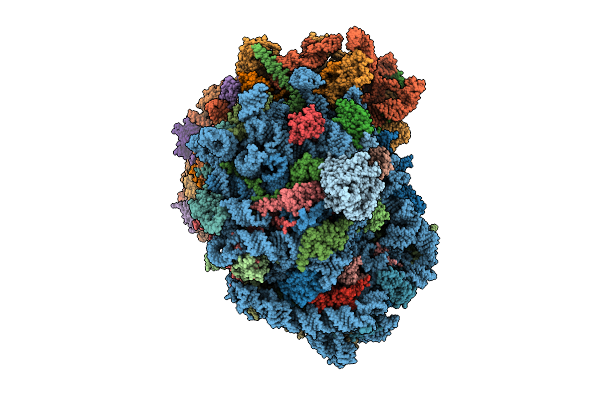 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
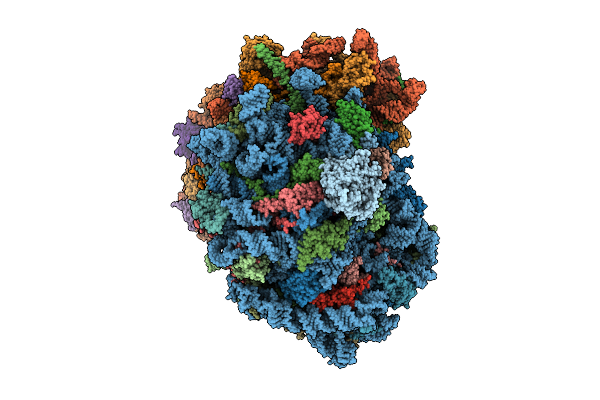 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
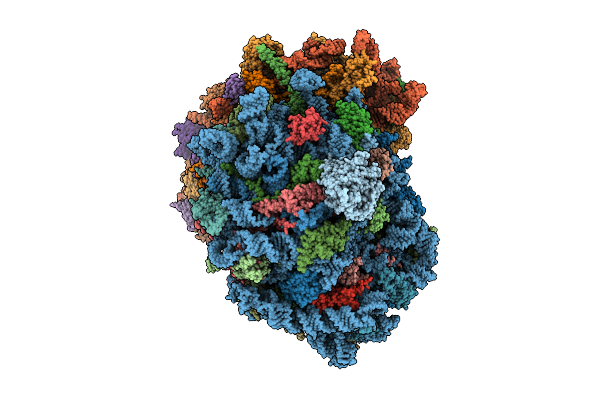 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
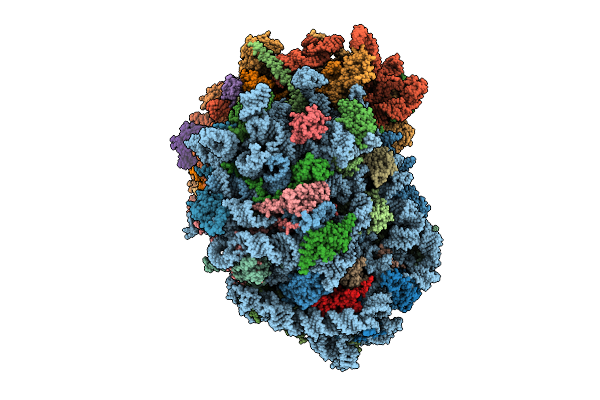 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: ZN, MG, SPM, SPD, 3HE, HMT |
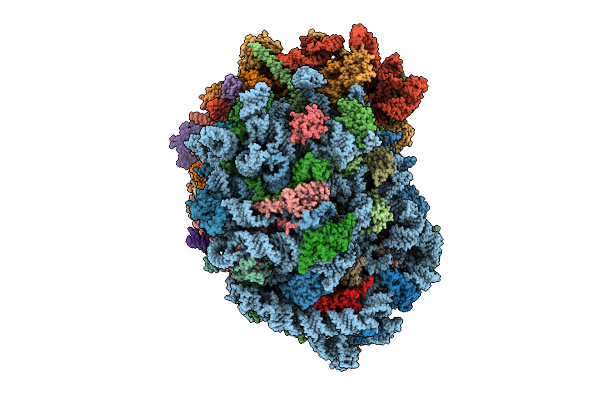 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, ZN, SPM, SPD, 3HE, HMT |
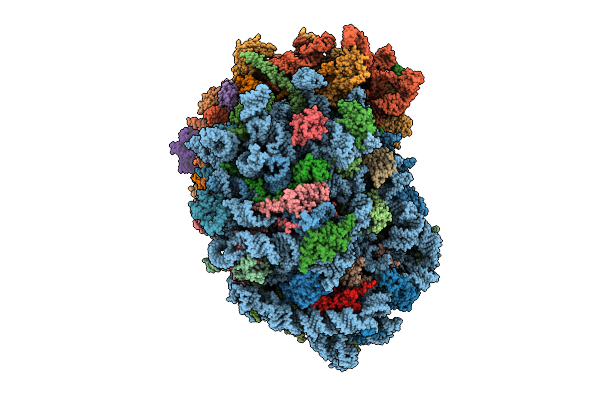 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, HMT, 3HE, ZN |
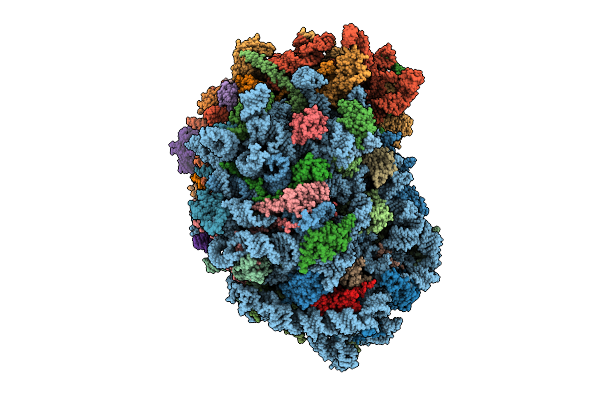 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, 3HE, HMT, ZN |
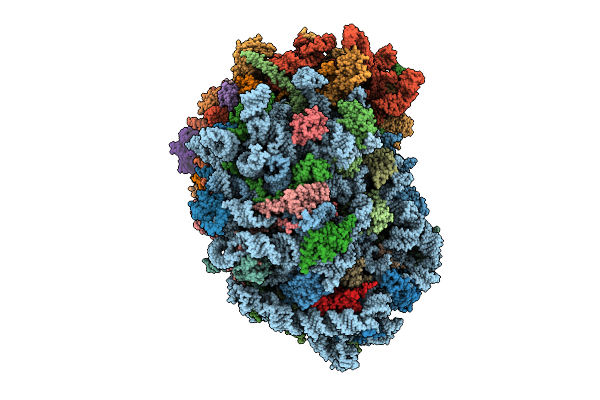 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, HMT, 3HE, ZN |
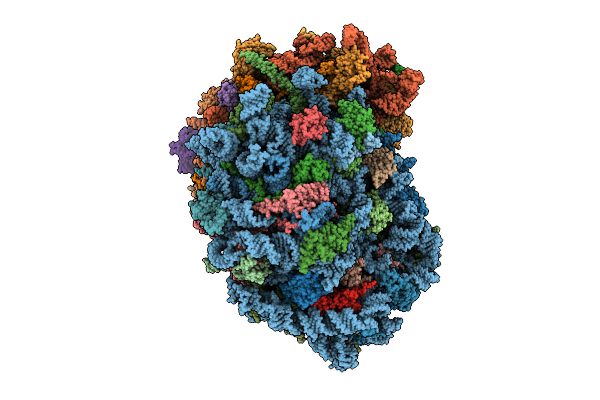 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
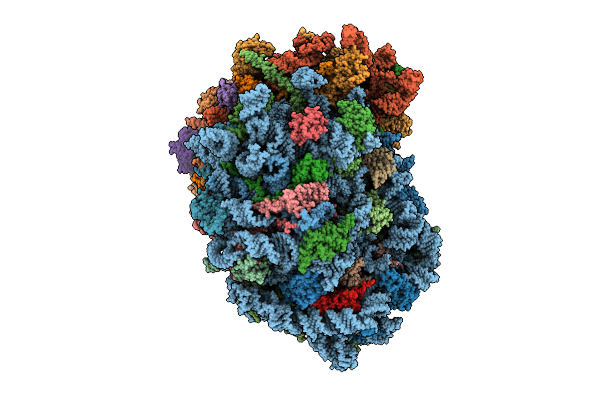 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
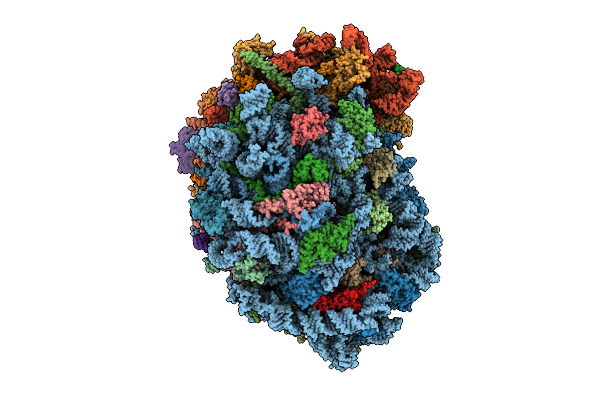 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
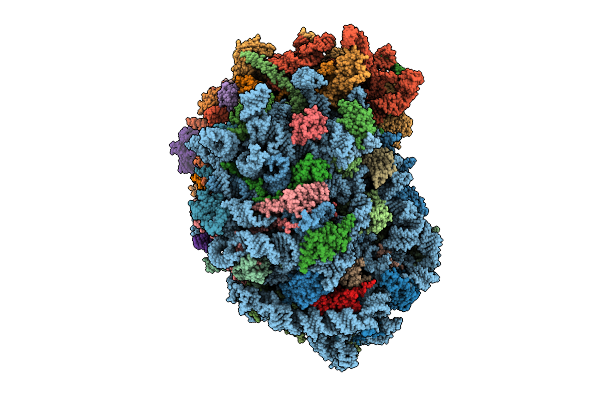 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
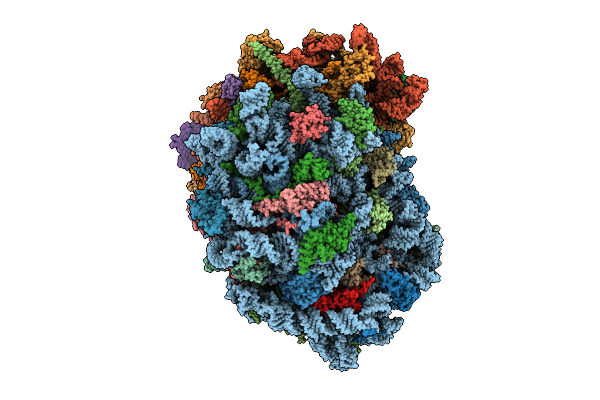 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, ZN, SPM, SPD |