Search Count: 294
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: CLR, T7M, ZKV |
 |
Organism: Methanocaldococcus jannaschii
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: METAL BINDING PROTEIN Ligands: SF4 |
 |
Organism: Litorilinea aerophila
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: YVF |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: YVF |
 |
Organism: Canis lupus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: A1AQX, NAG |
 |
Organism: Canis lupus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: MEMBRANE PROTEIN Ligands: NAG, MG, ATP |
 |
Crystal Structure Of Dna/Rna Duplex Obtained Using The Counter Diffusion Method On Earth (K Form)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-02-19 Classification: RNA/DNA Ligands: SPM |
 |
Crystal Structure Of Dna/Rna Duplex Obtained Using The Counter Diffusion Method In Space (K Form)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-02-19 Classification: RNA/DNA Ligands: SPM |
 |
Crystal Structure Of Dna/Rna Duplex Obtained Using The Counter Diffusion Method On Earth (Na Form)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-19 Classification: DNA/RNA Ligands: SPM |
 |
Crystal Structure Of Dna/Rna Duplex Obtained Using The Counter Diffusion Method In Space (Na Form)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-19 Classification: DNA/RNA Ligands: SPM |
 |
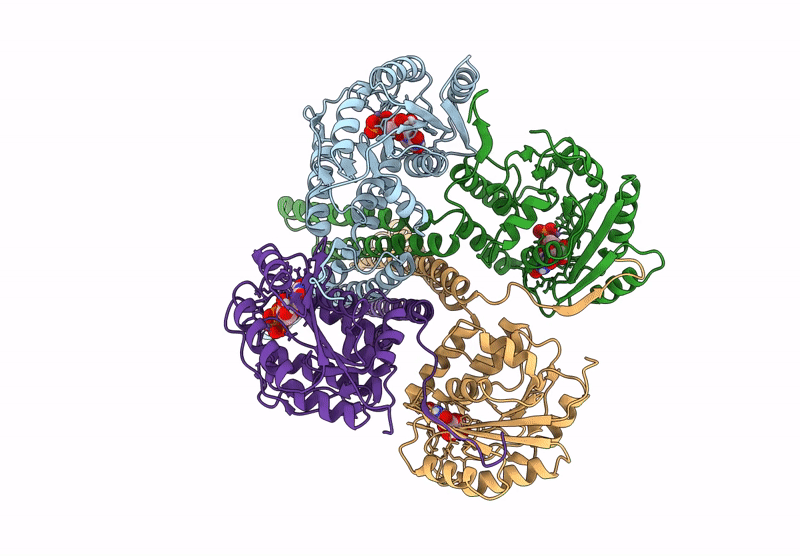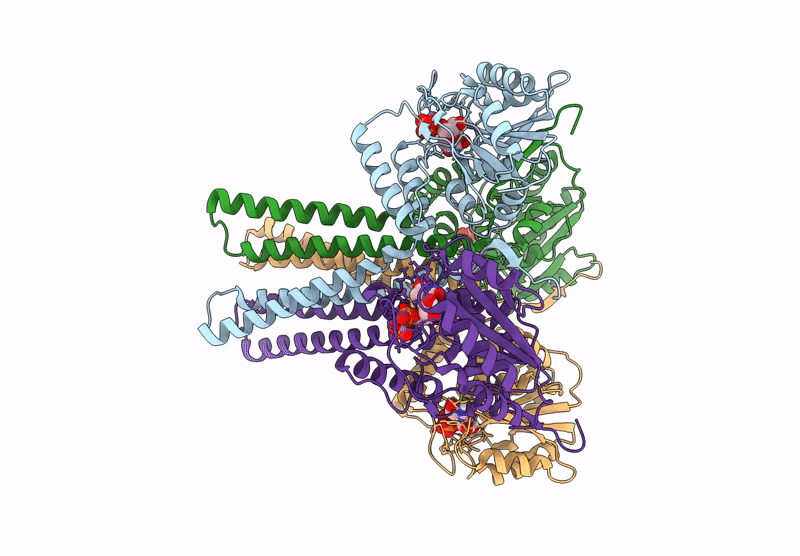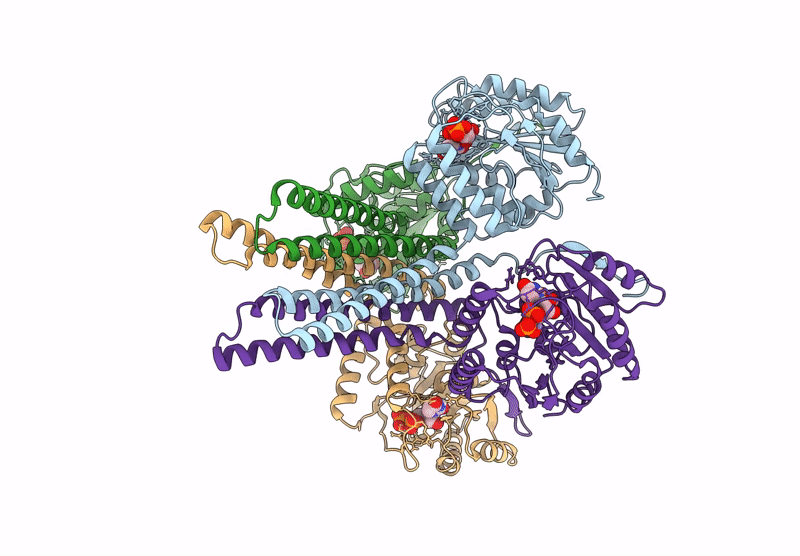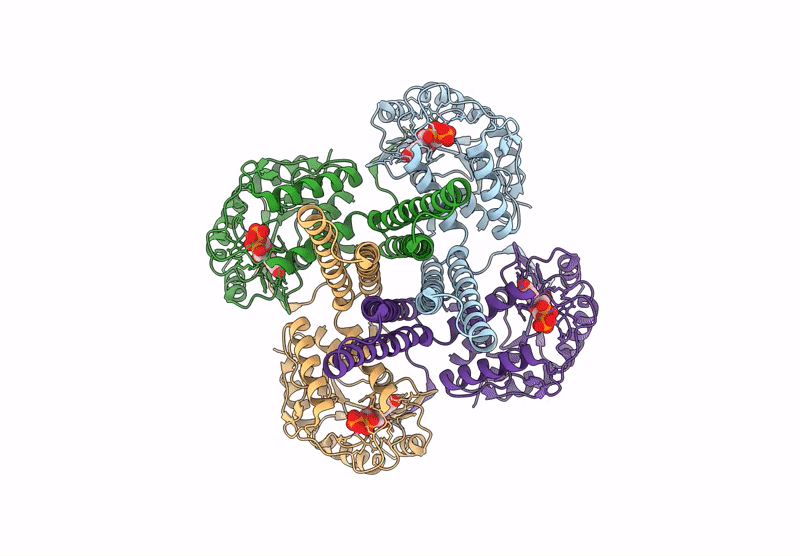
Cryo-Em Structure Of The Glycosyltransferase Arnc From Salmonella Enterica In The Apo State Determined On Talos Arctica Microscope
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE |
 |
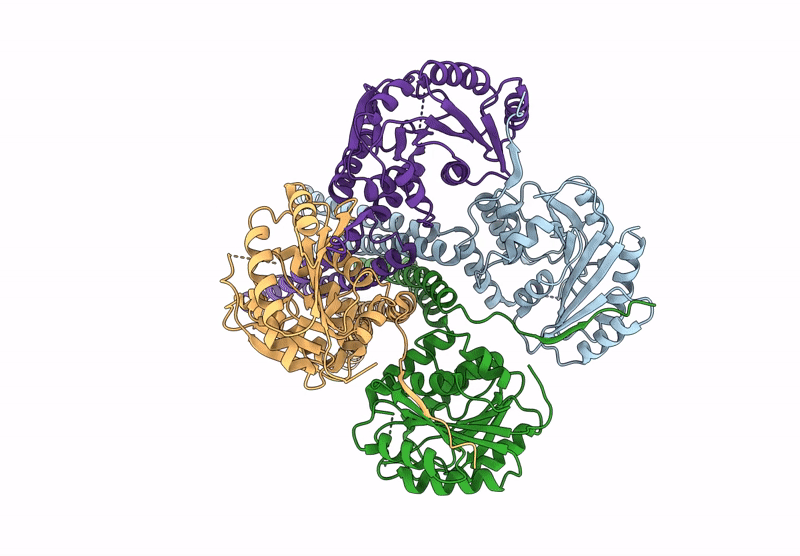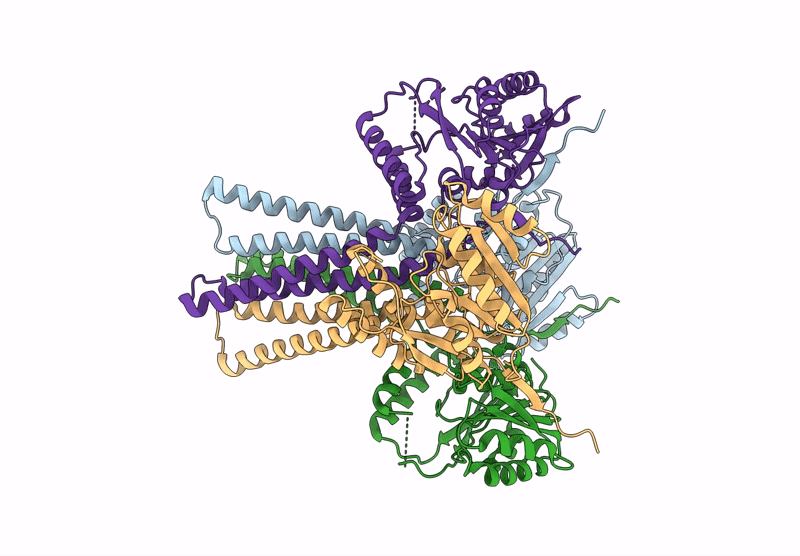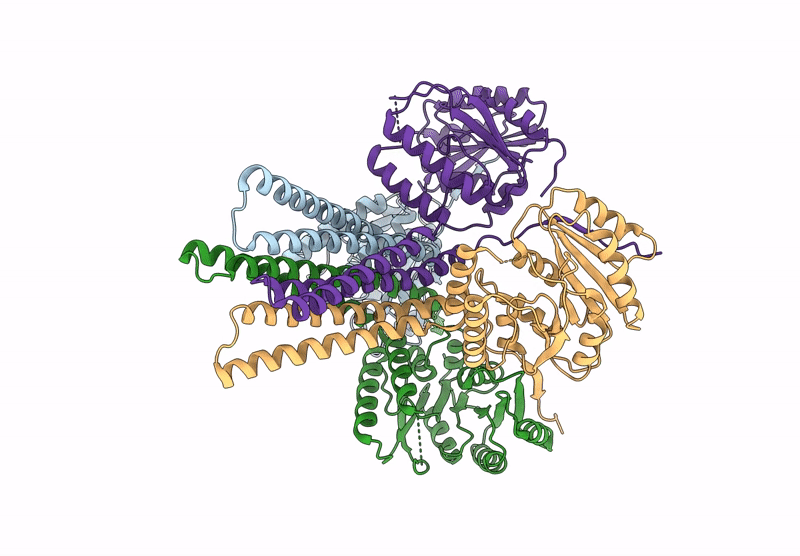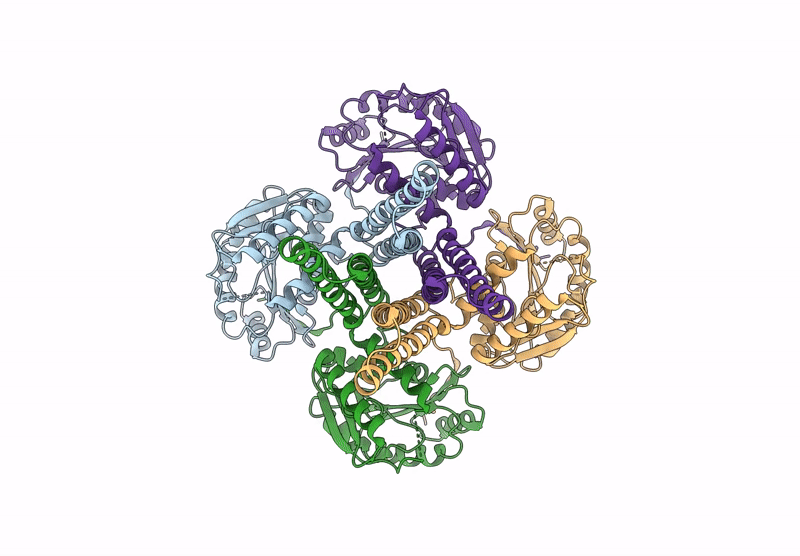
Cryo-Em Structure Of The Glycosyltransferase Arnc From Salmonella Enterica In The Udp-Bound State Determined On Talos Arctica Microscope
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: UDP, MN |
 |
Cryo-Em Structure Of The Glycosyltransferase Arnc From Salmonella Enterica In The Apo State Determined On Krios Microscope
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE |
 |
Organism: Neobacillus niacini
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: FAD, DR9 |
 |
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, DR9 |
 |
Organism: Neobacillus niacini
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: CYTOSOLIC PROTEIN Ligands: FAD, WTQ, DR9 |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-09-04 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-08-14 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-14 Classification: HYDROLASE Ligands: MER, MG, ZN |

