Search Count: 25
 |
Lytic Transglycosylase Mltd Of Pseudomonas Aeruginosa Bound To The Natural Product Bulgecin A
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-17 Classification: LYASE Ligands: BLG, ZN |
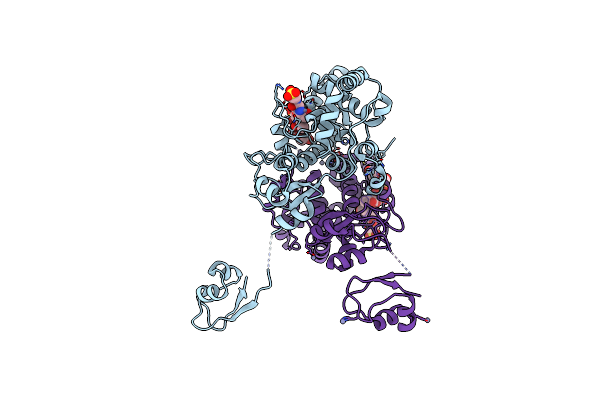 |
Lytic Transglycosylase Mltd Of Pseudomonas Aeruginosa Bound To The Natural Product Bulgecin A, With Two Lysm Domains
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-17 Classification: LYASE Ligands: PEG, BLG, ZN |
 |
Lytic Transglycosylase Mltd Of Pseudomonas Aeruginosa In A Ternary Complex Bound To Bulgecin A And Chito-Tetraose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-04-17 Classification: LYASE Ligands: BLG, ZN |
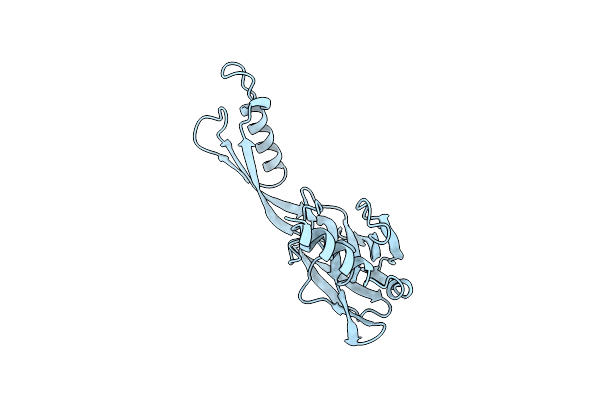 |
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-02-02 Classification: IMMUNE SYSTEM |
 |
Organism: Schistosoma mansoni
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2020-09-30 Classification: ALLERGEN |
 |
Organism: Alicyclobacillus tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-07-29 Classification: HYDROLASE Ligands: B3P, EDO, NA, IOD |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: SO4, MG |
 |
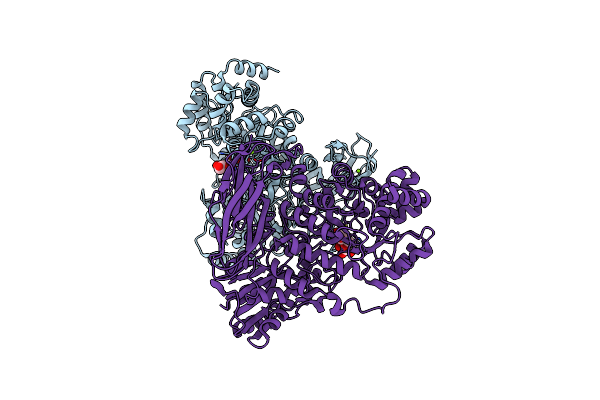
The Crystal Structure Of Glycoside Hydrolase Bglx From P. Aeruginosa In Complex With 1-Deoxynojirimycin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, NOJ, MG |
 |
The Crystal Structure The Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Two Glucose Molecules
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
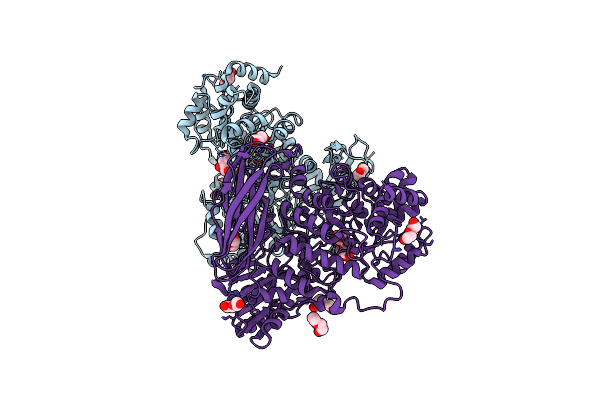
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Glucose
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Cellobiose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PEG, PGE, MG |
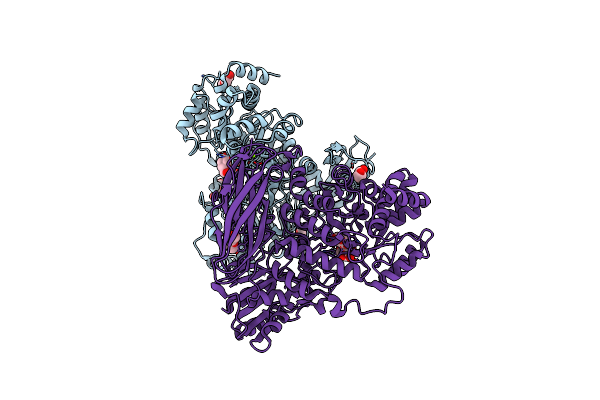 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Lactose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PEG, MG, PGE |
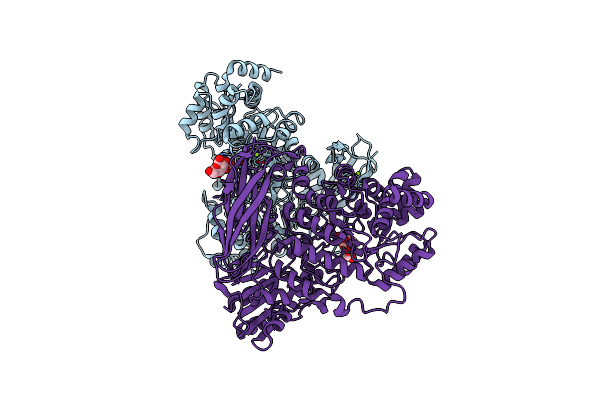 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant From P. Aeruginosa In Complex With Laminaritriose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: MG |
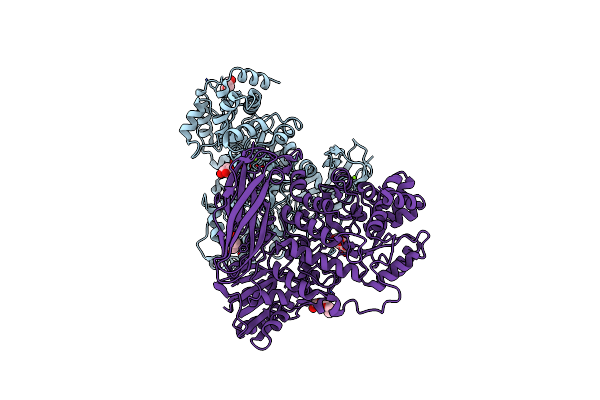 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Xylotriose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, PEG, MG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2019-11-13 Classification: CELL CYCLE |
 |
Crystal Structure Of Rlpa Spor Domain From Pseudomonas Aeruginosa In Complex With Denuded Glycan Obtained By Soaking
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-11-13 Classification: CELL CYCLE |
 |
Crystal Structure Of Rlpa Spor Domain From Pseudomonas Aeruginosa In Complex With Nuded Glycan Obtained By Co-Crystallization
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-11-13 Classification: CELL CYCLE |
 |
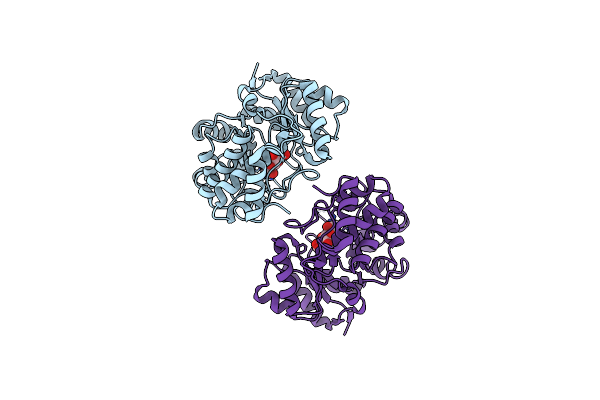
Crystal Structure Of Rlpa Spor Domain From Pseudomonas Aeruginosa In Complex With Denuded Glycan Ended In Anhnam
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-13 Classification: CELL CYCLE Ligands: AH0 |
 |
Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-Acetylglucosamine
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-05-17 Classification: HYDROLASE Ligands: NAG |
 |
Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-Acetylglucosamine And L-Ala-1,6-Anhydromurnac
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2017-05-17 Classification: HYDROLASE Ligands: NAG, 89A, PEG |

