Search Count: 243
 |
Functional And Structural Insights Into The Unusual Oxyanion Hole-Like Geometry In Macrolactin Acyltransferase Selective For Dicarboxylic Acyl Donors
Organism: Jeotgalibacillus marinus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: SO4 |
 |
Functional And Structural Insights Into The Unusual Oxyanion Hole-Like Geometry In Macrolactin Acyltransferase Selective For Dicarboxylic Acyl Donors
Organism: Jeotgalibacillus marinus
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: D9L |
 |
Functional And Structural Insights Into The Unusual Oxyanion Hole-Like Geometry In Macrolactin Acyltransferase Selective For Dicarboxylic Acyl Donors
Organism: Jeotgalibacillus marinus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: SO4, D9F |
 |
Functional And Structural Insights Into The Unusual Oxyanion Hole-Like Geometry In Macrolactin Acyltransferase Selective For Dicarboxylic Acyl Donors
Organism: Jeotgalibacillus marinus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: D9F |
 |
Functional And Structural Insights Into The Unusual Oxyanion Hole-Like Geometry In Macrolactin Acyltransferase Selective For Dicarboxylic Acyl Donors
Organism: Jeotgalibacillus marinus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: D9O |
 |
Functional And Structural Insights Into The Unusual Oxyanion Hole-Like Geometry In Macrolactin Acyltransferase Selective For Dicarboxylic Acyl Donors
Organism: Jeotgalibacillus marinus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-07-29 Classification: TRANSFERASE Ligands: SIN |
 |
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-29 Classification: LIPID BINDING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-07-29 Classification: CELL ADHESION |
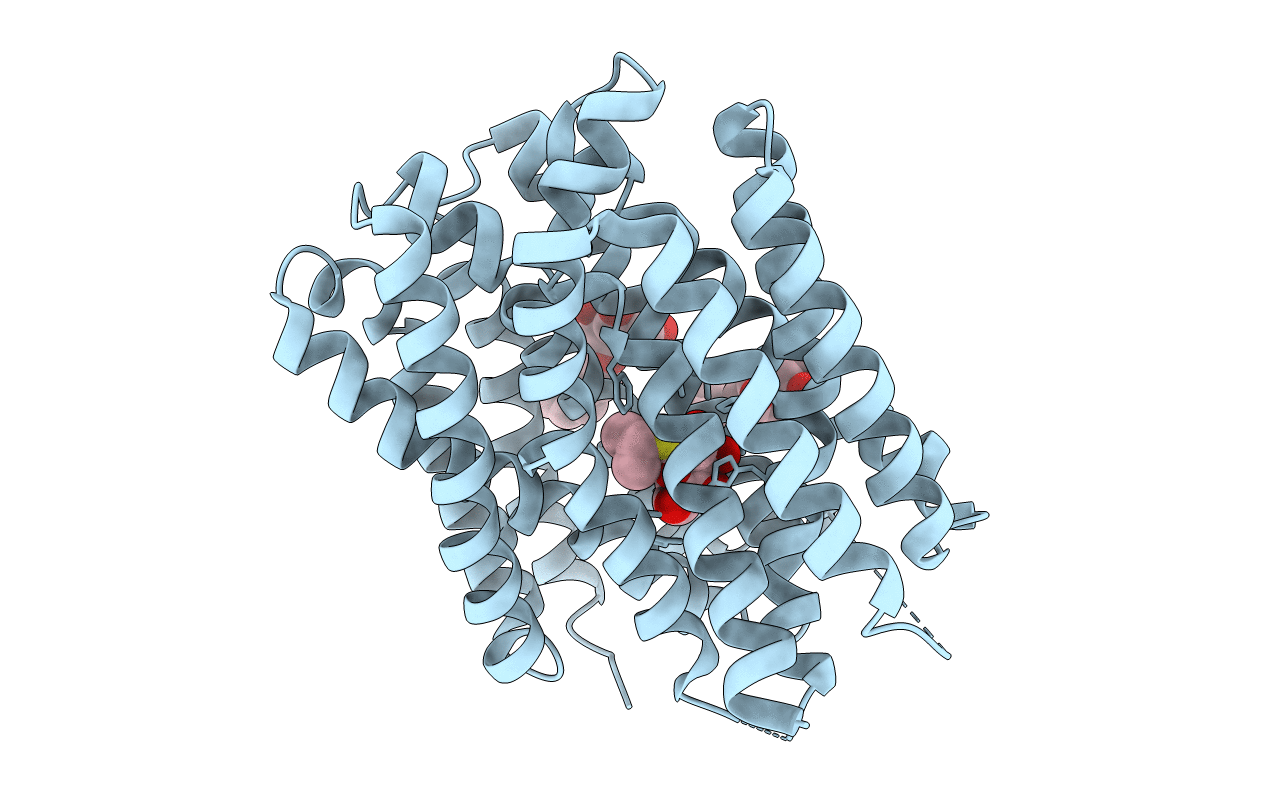 |
Crystal Structure Of Drug:Proton Antiporter-1 (Dha1) Family Sotb, In The Inward-Occluded Conformation
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2020-07-29 Classification: PROTEIN TRANSPORT Ligands: BNG, IPT |
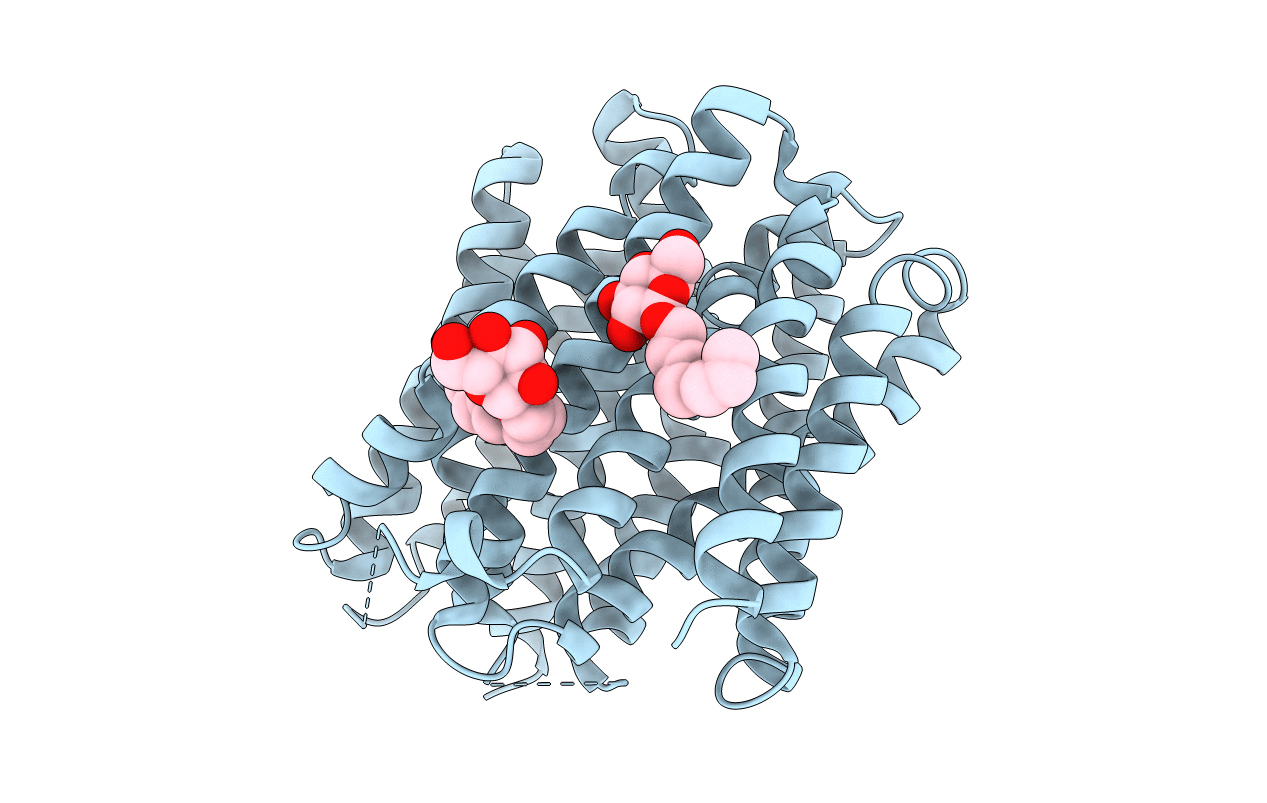 |
Crystal Structure Of Drug:Proton Antiporter-1 (Dha1) Family Sotb, In The Inward Open Conformation
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2020-07-29 Classification: PROTEIN TRANSPORT Ligands: BNG |
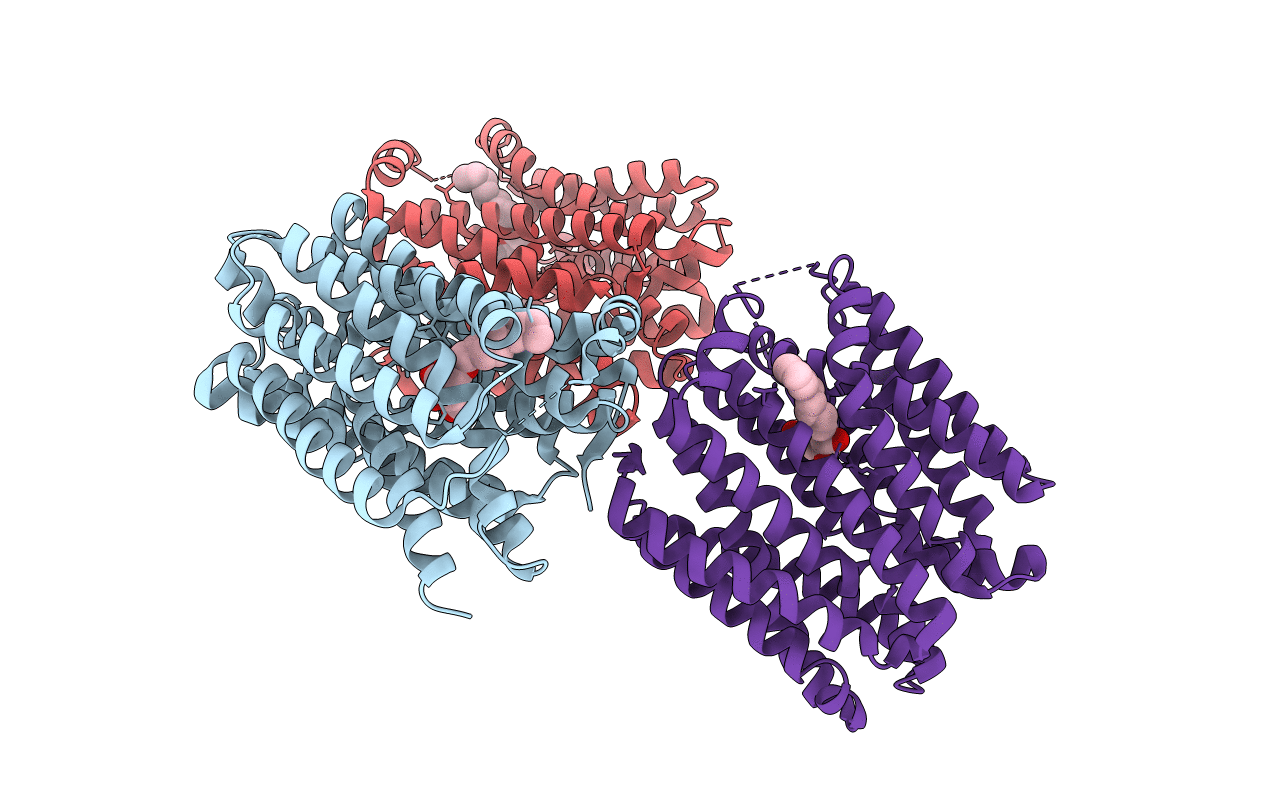 |
Crystal Structure Of Drug:Proton Antiporter-1 (Dha1) Family Sotb, In The Inward Open Conformation (H115A Mutant)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-07-29 Classification: TRANSPORT PROTEIN Ligands: BNG |
 |
Crystal Structure Of Drug:Proton Antiporter-1 (Dha1) Family Sotb, In The Inward Conformation (H115N Mutant)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-07-29 Classification: TRANSPORT PROTEIN Ligands: BNG |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2020-07-29 Classification: LIGASE Ligands: DKR, SO4 |
 |
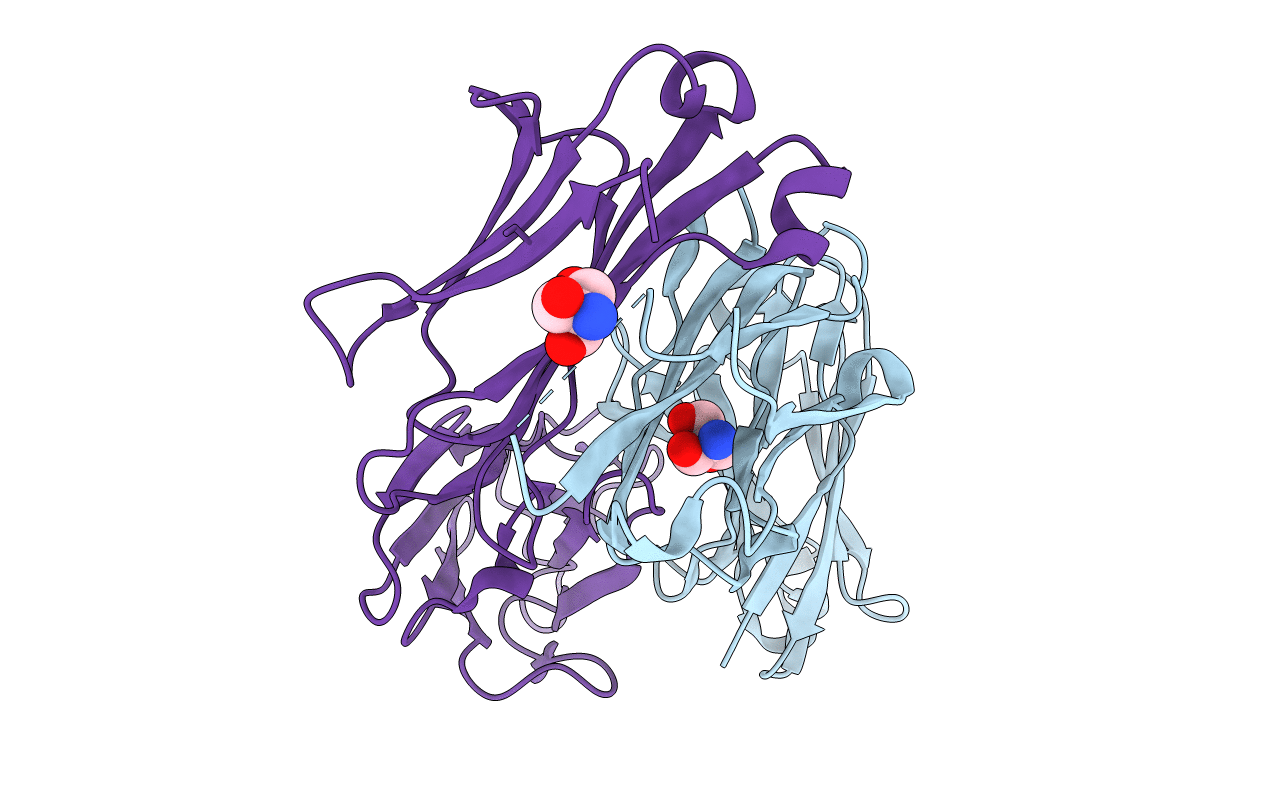
100K X-Ray Structure Of Hiv-1 Protease Triple Mutant (V32I,I47V,V82I) With Tetrahedral Intermediate Mimic Kvs-1
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: KVS |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-07-29 Classification: IMMUNE SYSTEM Ligands: TRS |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-07-29 Classification: IMMUNE SYSTEM Ligands: A2G |
 |
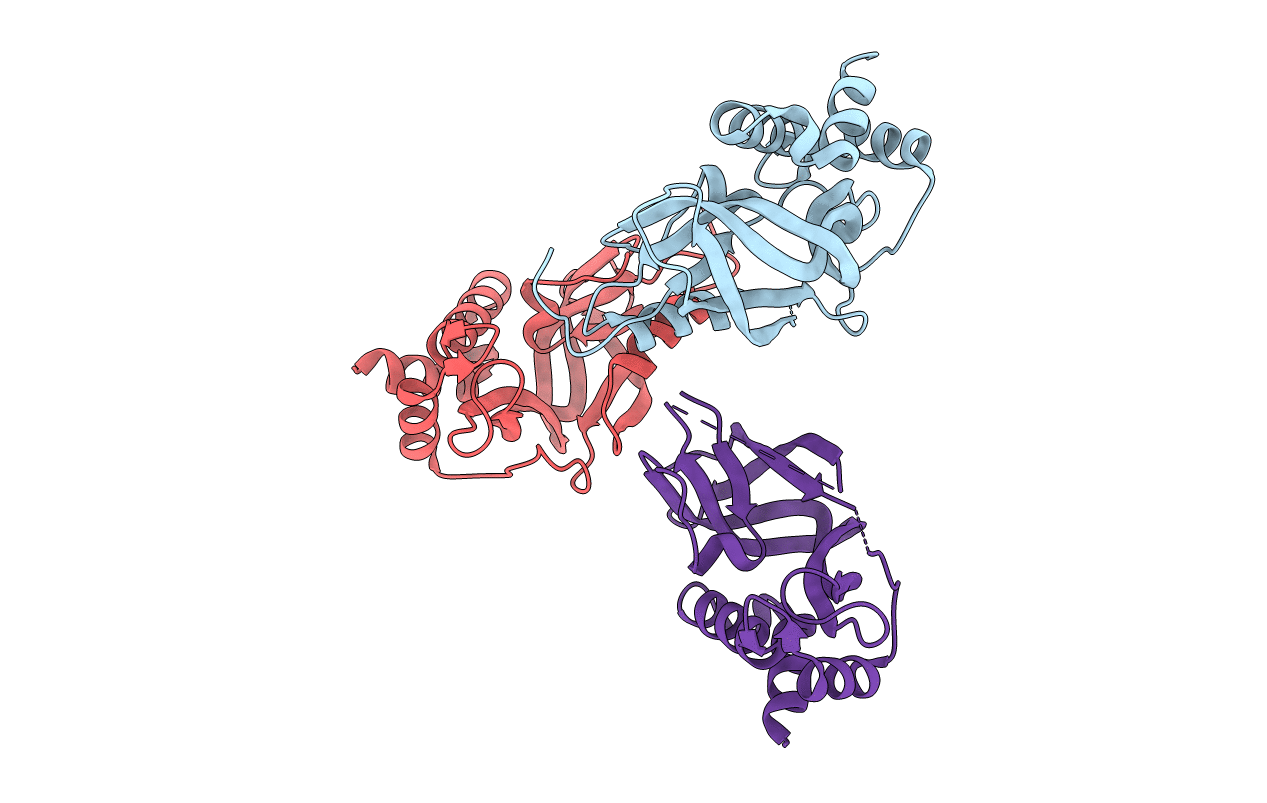
Crystal Structural Of Active-Site Human Glutathione-Specific Gamma-Glutamylcyclotransferase 2(Chac2) Mutant With Glutamate 74 Replaced By Glutamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-07-29 Classification: TRANSFERASE |
 |
Crystal Structural Of Human Glutathione-Specific Gamma-Glutamylcyclotransferase 2 (Chac2)Mutant With Glutamate 83 Replaced By Glutamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-07-29 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-07-29 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2020-07-29 Classification: STRUCTURAL PROTEIN |

