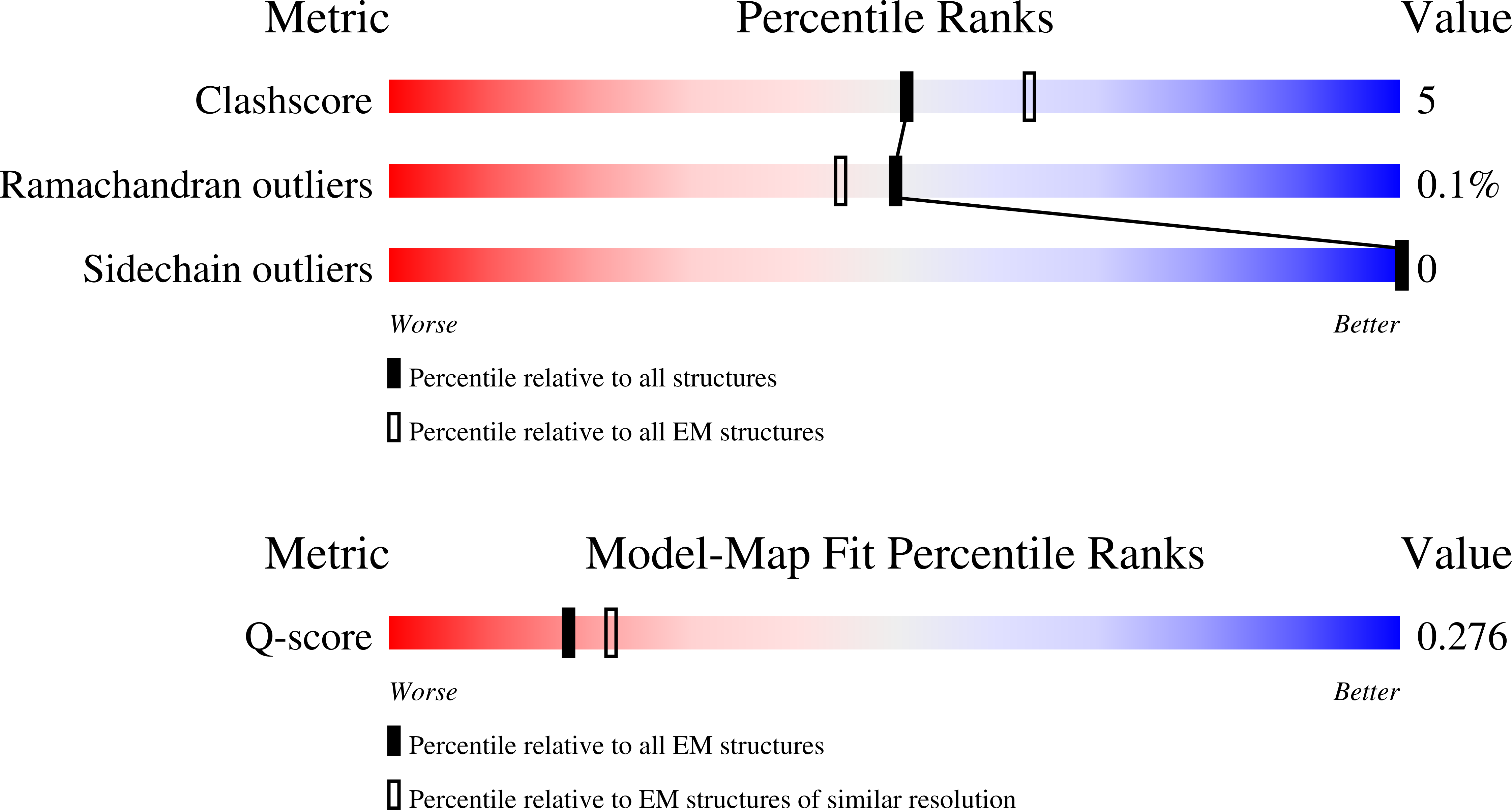
Deposition Date
2025-09-29
Release Date
2025-11-19
Last Version Date
2026-01-28
Entry Detail
PDB ID:
9YGY
Keywords:
Title:
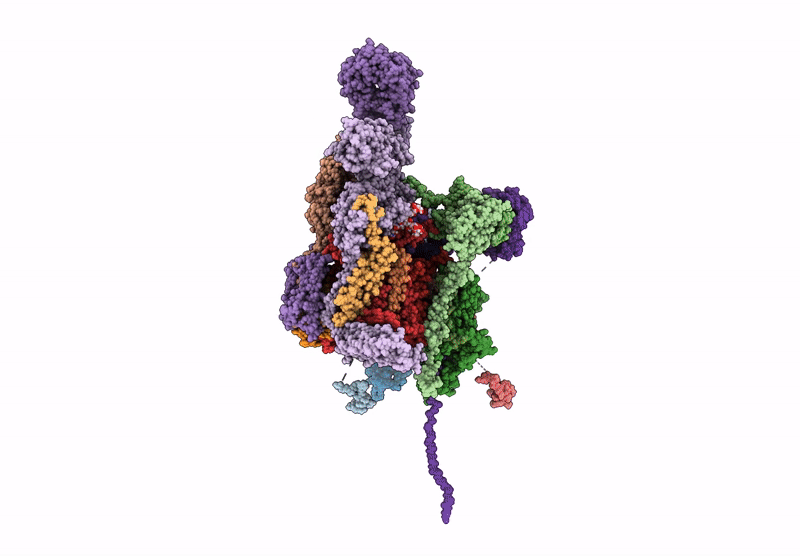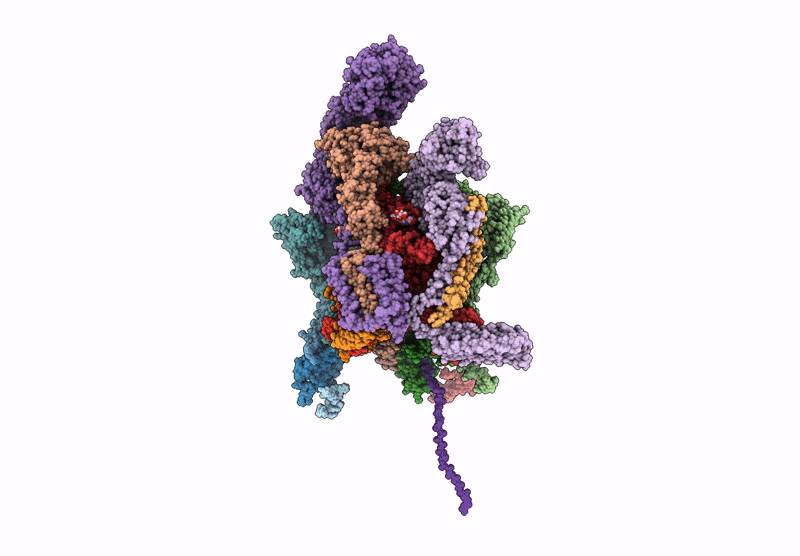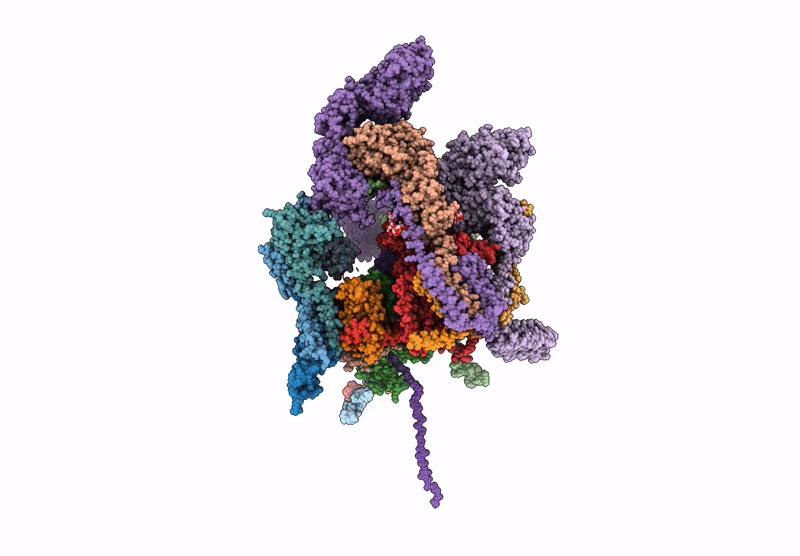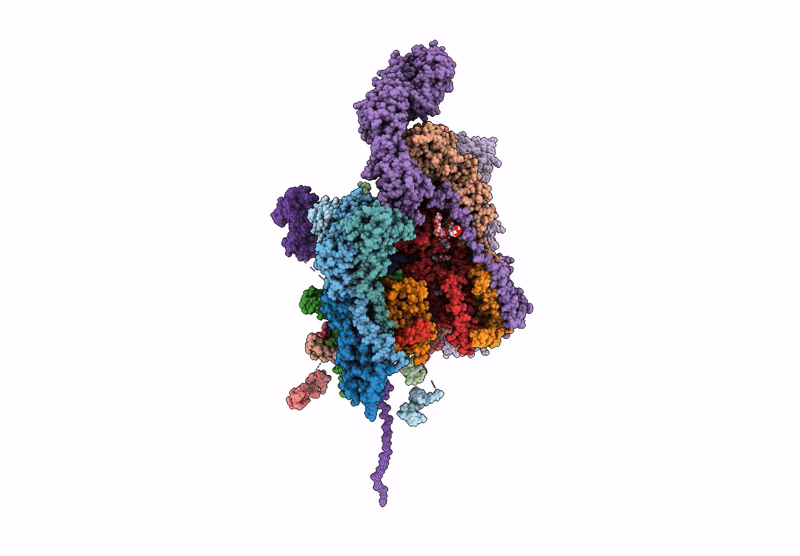
Structure of a GRP94 folding intermediate engaged with a CCDC134- and FKBP11-bound secretory translocon
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
Resolution:
4.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE