Deposition Date
2025-09-15
Release Date
2025-11-26
Last Version Date
2025-12-24
Entry Detail
PDB ID:
9WSX
Keywords:
Title:
Cryo-EM structure of endomorphin-1-muOR-arrestin2-Fab30 complex
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Homo sapiens (Taxon ID: 9606)
Bos taurus (Taxon ID: 9913)
synthetic construct (Taxon ID: 32630)
Homo sapiens (Taxon ID: 9606)
Bos taurus (Taxon ID: 9913)
synthetic construct (Taxon ID: 32630)
Expression System(s):
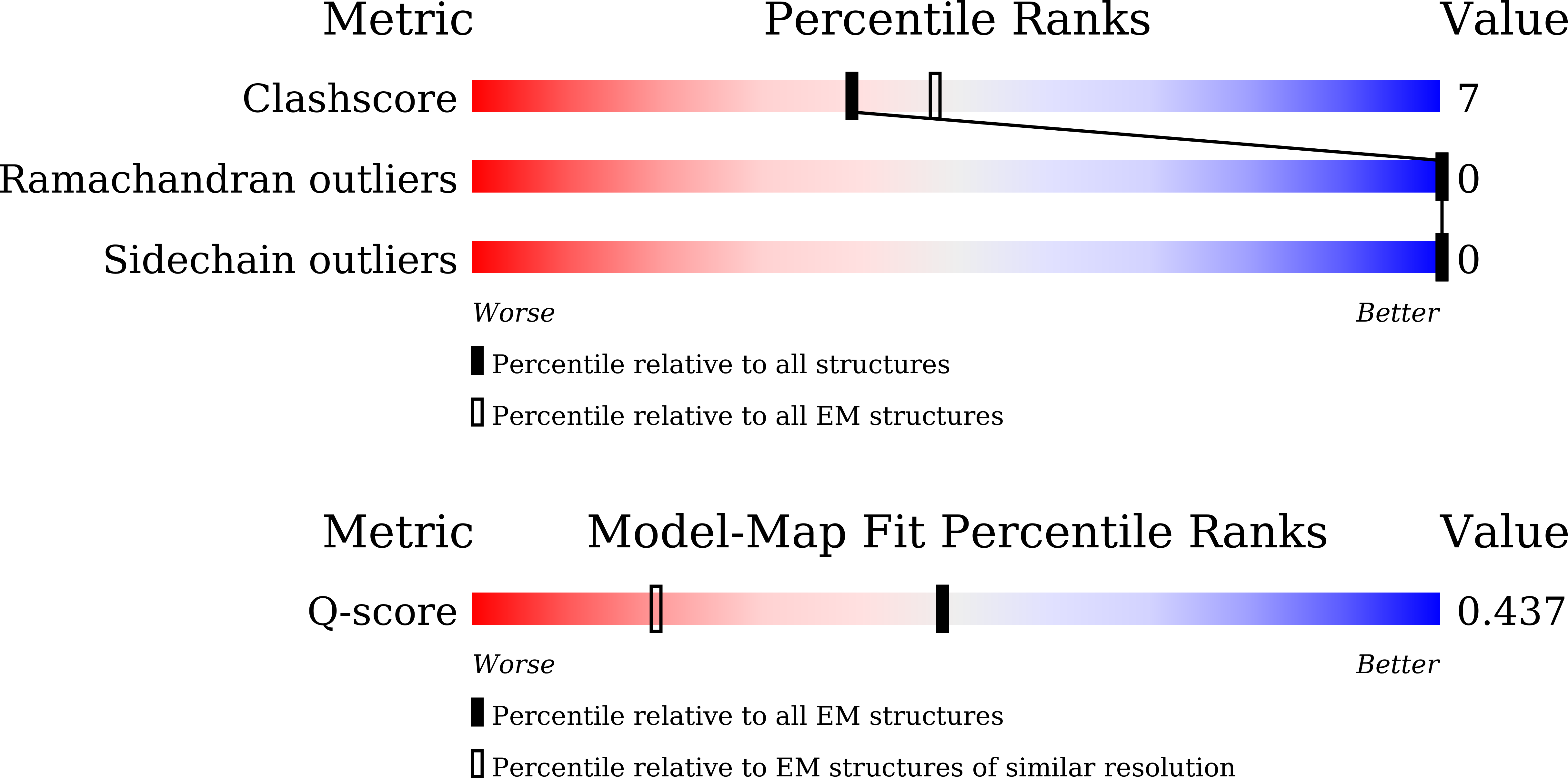
Method Details:
Experimental Method:
Resolution:
2.80 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE