Deposition Date
2025-09-03
Release Date
2025-12-24
Last Version Date
2025-12-24
Entry Detail
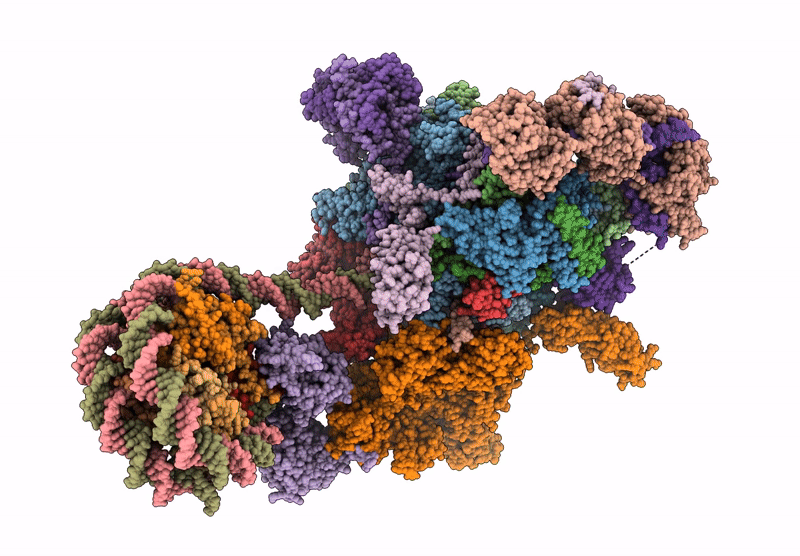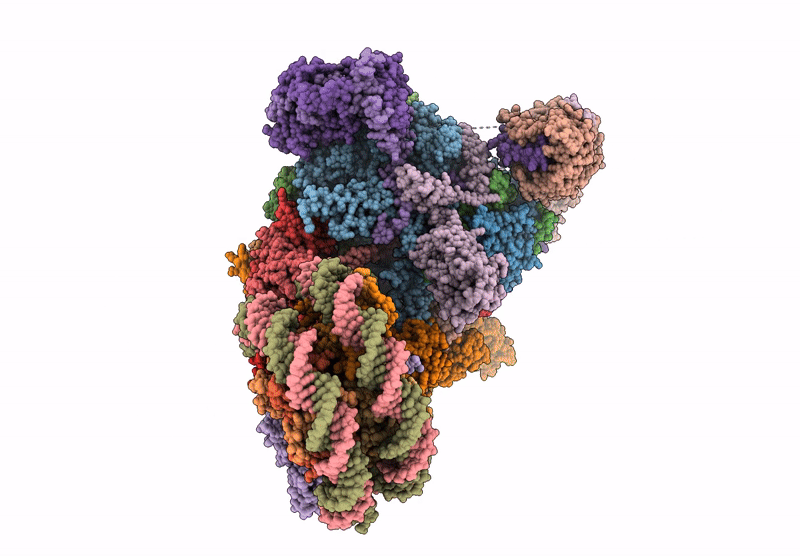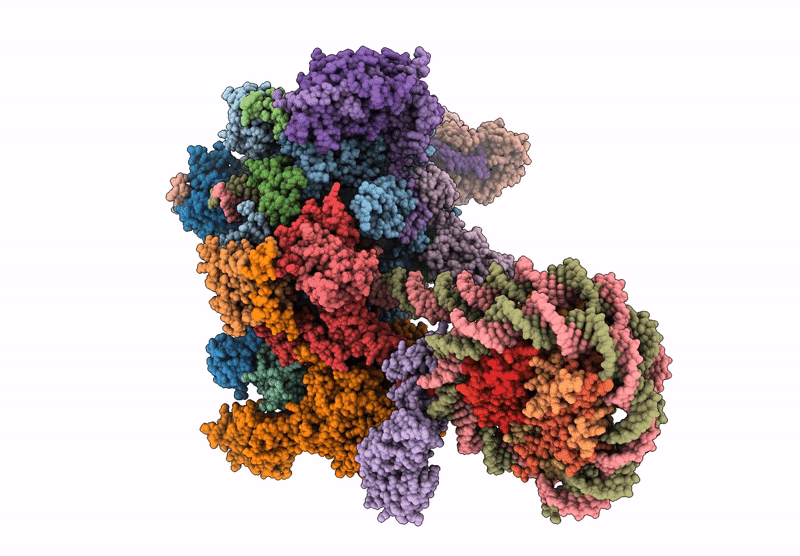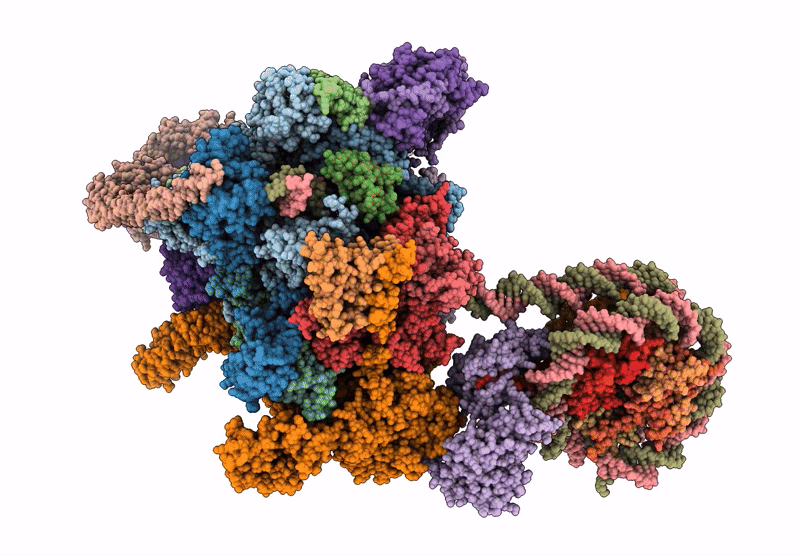
PDB ID:
9WMV
Keywords:
Title:
Co-transcriptional histone H3K36 methylation complex containing RNA polymerase II elongation complex, Set2, and the upstream nucleosome. (temp130, type B)
Biological Source:
Source Organism:
Komagataella phaffii GS115 (Taxon ID: 644223)
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
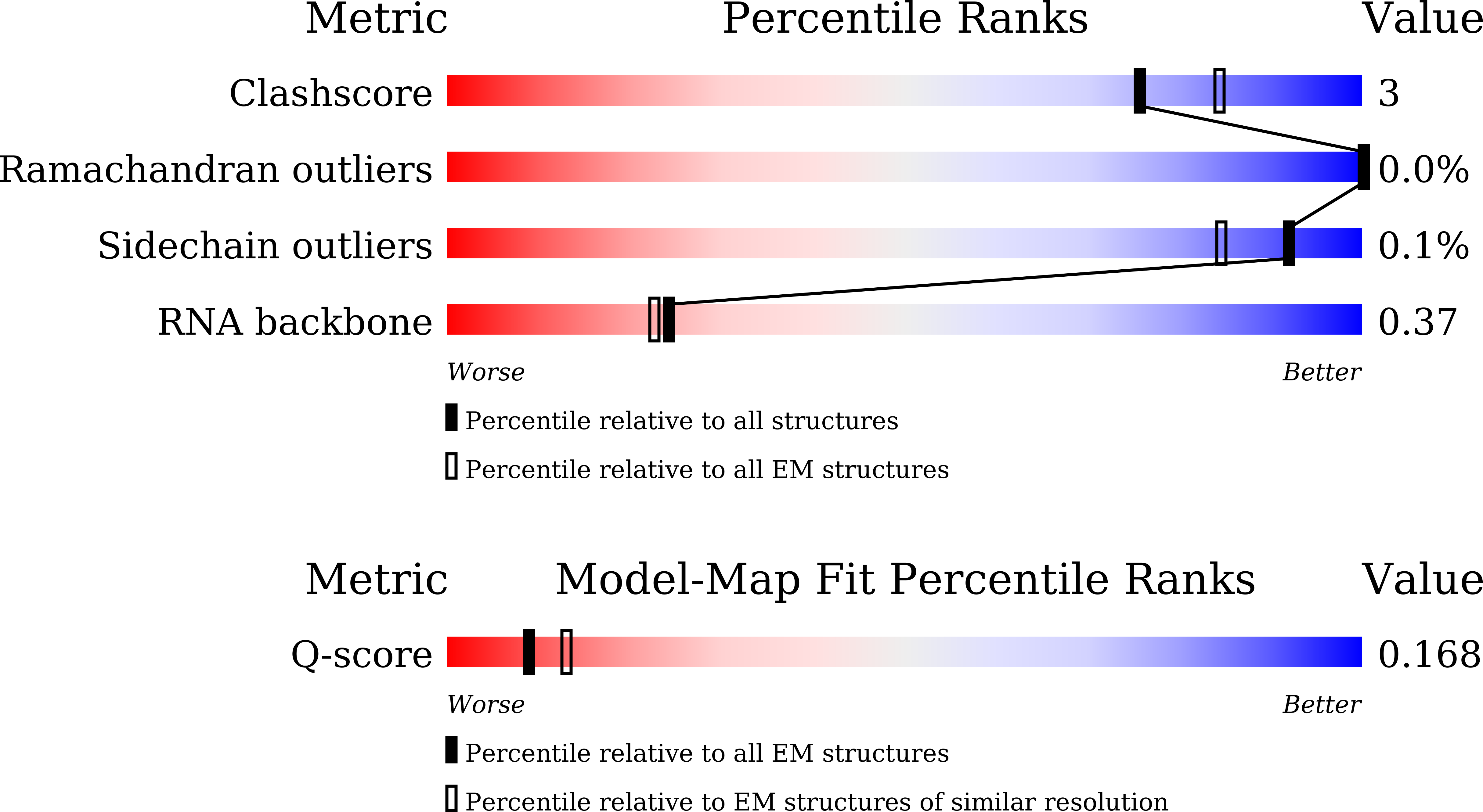
Resolution:
4.66 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE