Abstact
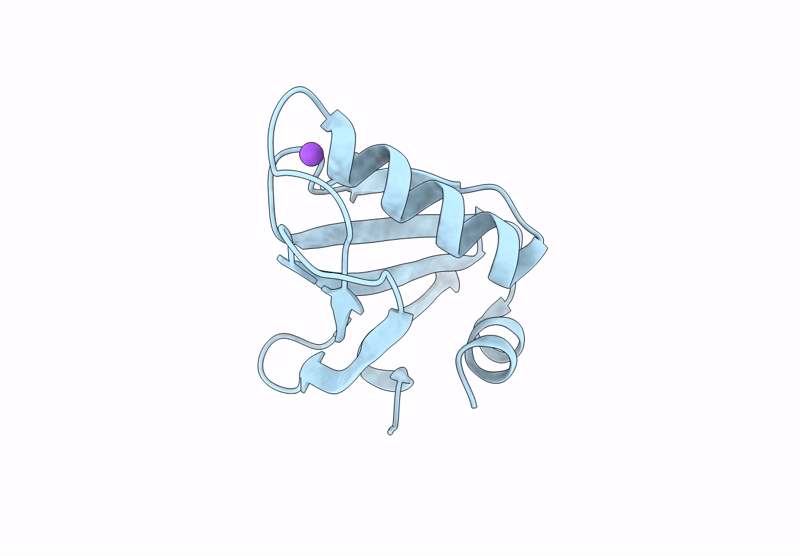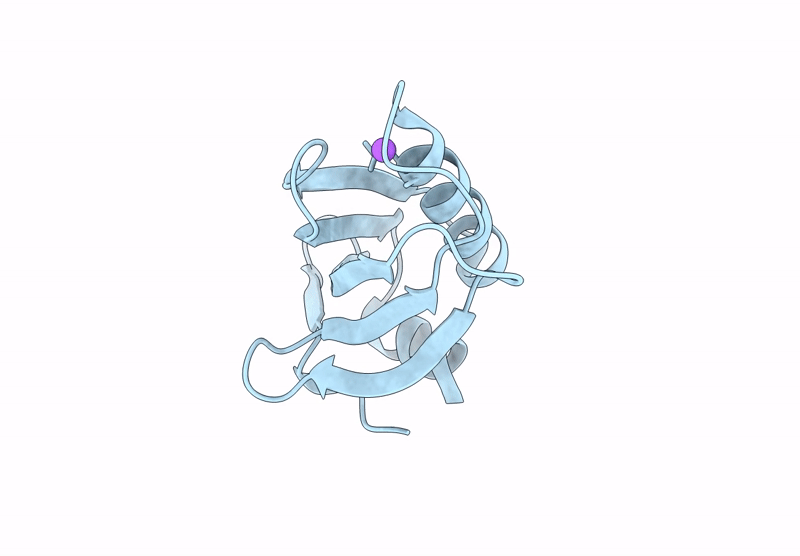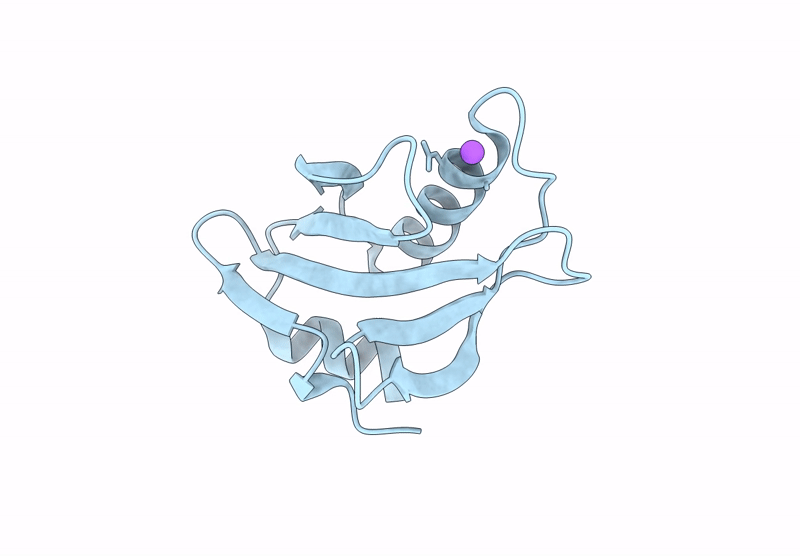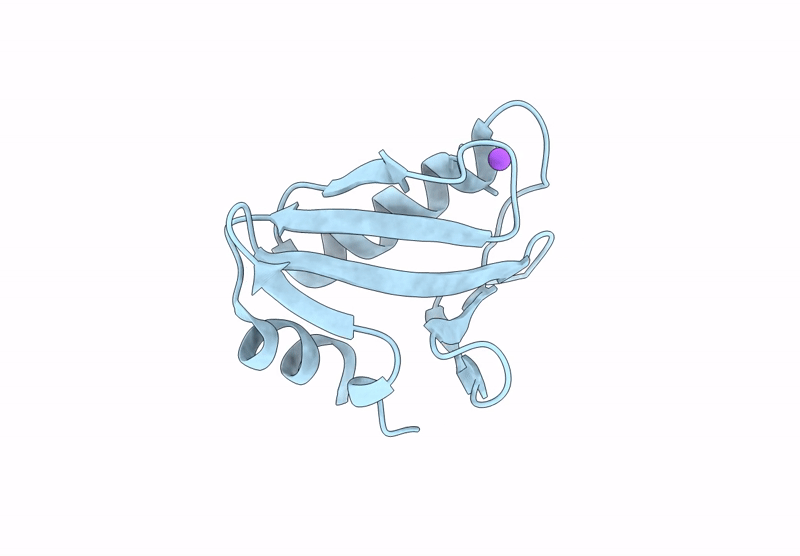
Proteins crystallized under varied conditions typically exhibit nearly identical overall structures, with deviations confined to flexible loops or side-chain orientations. In this study, we report the extraordinary crystallization properties of PF1765 from Pyrococcus furiosus, which crystallized from the same batch of protein preparation in 104 of 192 different crystallization conditions. This yielded ten high-resolution structures (1.1-1.5 Å) across two space groups: seven in the orthorhombic space group P212121 (including one previously reported, PDB entry 9unt) and three in the tetragonal P41212. Despite large variations in pH, salt and precipitant, all structures were nearly identical, with pairwise Cα r.m.s.d.s of 0.06-0.33 Å. Structures within the same space group were indistinguishable, with pairwise Cα r.m.s.d.s of 0.06-0.09 and 0.09-0.14 Å for the tetragonal and orthorhombic space groups, respectively. These results confirm that the overall structure remained unaffected by the large chemical variability during crystallization. Consistently, major crystal contacts were conserved across the two space groups, while hydration mapping identified six conserved waters across all of the structures. Interestingly, rotameric differences were observed between space groups, where residues Ser6, Glu7, Pro17, Asn18, Pro41, Pro42, Val43 and Arg72 adopt distinct conformations reflecting lattice-specific packing. Collectively, PF1765 emerges as a hyper-crystallizable protein that provides a consistent framework for analyzing lattice-dependent microheterogeneity, packing and hydration-site conservation at atomic resolution. Its compact, rigid single-domain structure and reproducible crystallization behavior indicate potential use as a fusion domain to aid the crystallization of membrane proteins or complexes by promoting ordered lattice formation. However, this study does not examine crystal nucleation or growth kinetics under varying conditions, which remain important directions for future investigation.