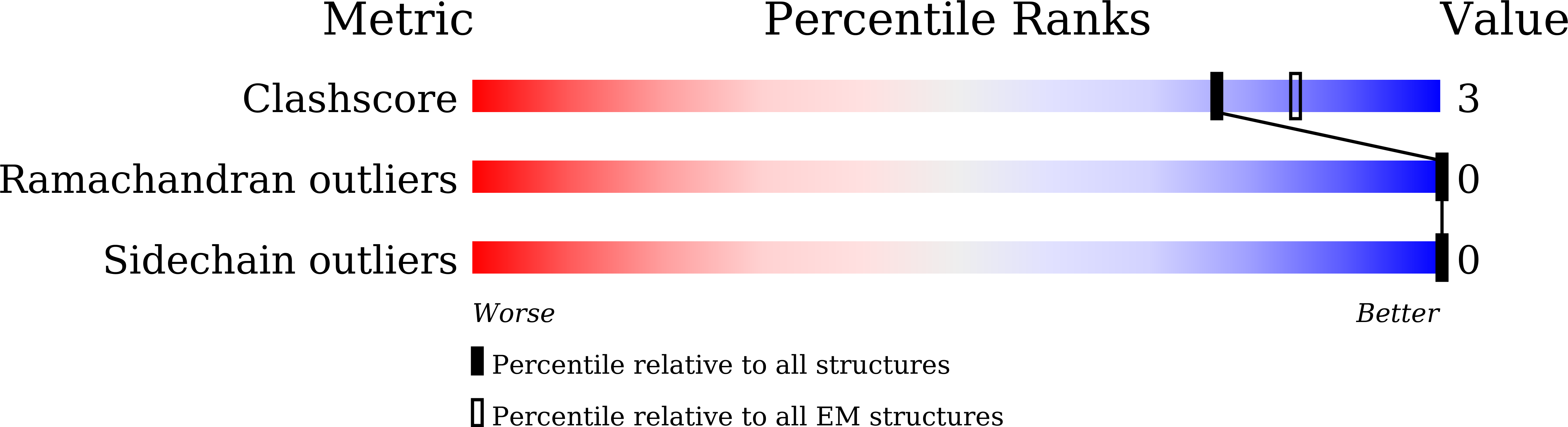
Deposition Date
2025-06-09
Release Date
2025-08-27
Last Version Date
2025-11-26
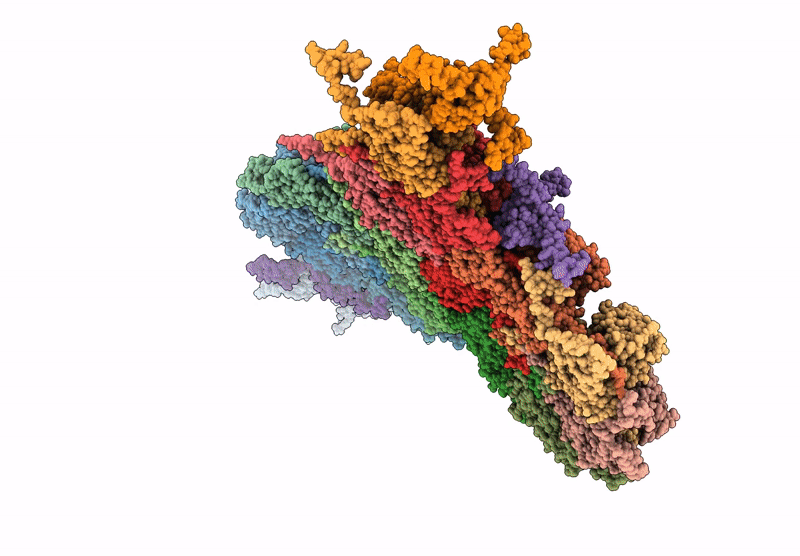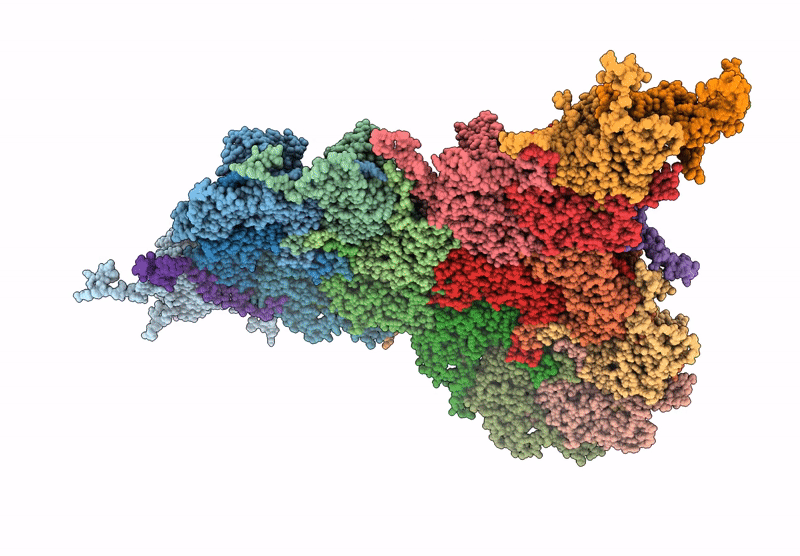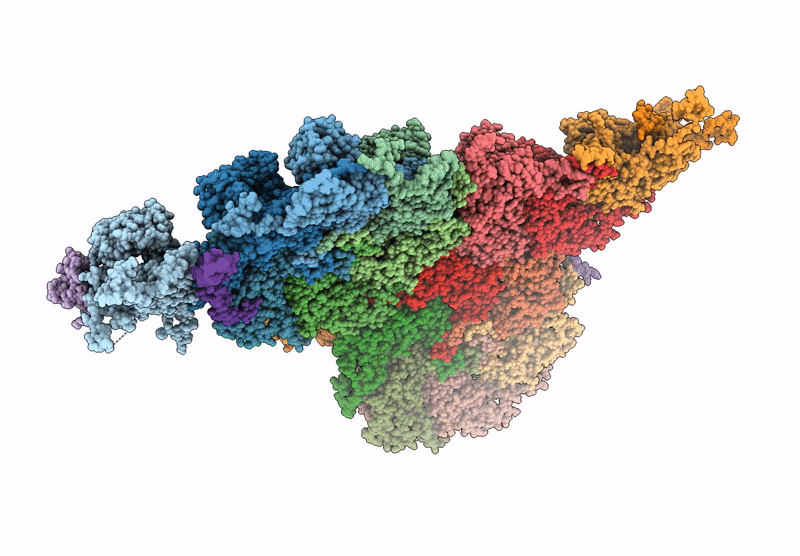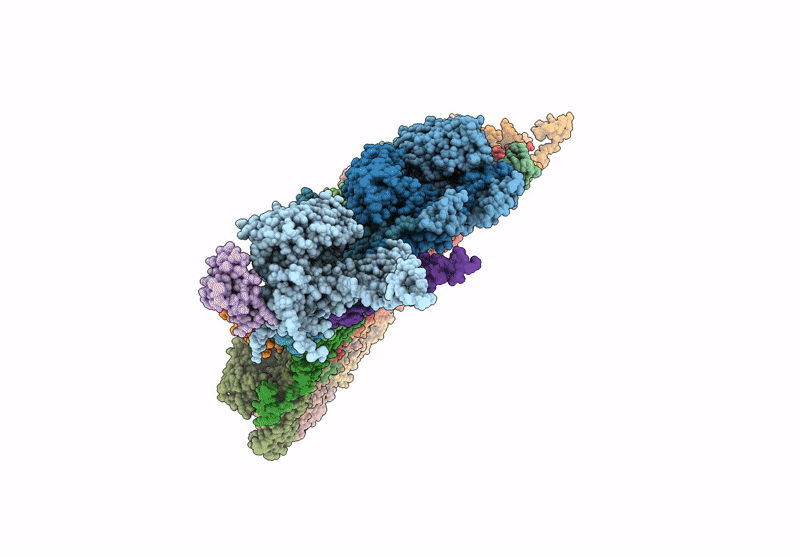
Method Details:
Experimental Method:
Resolution:
3.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE