Deposition Date
2025-05-22
Release Date
2025-10-08
Last Version Date
2025-11-26
Entry Detail
PDB ID:
9V44
Keywords:
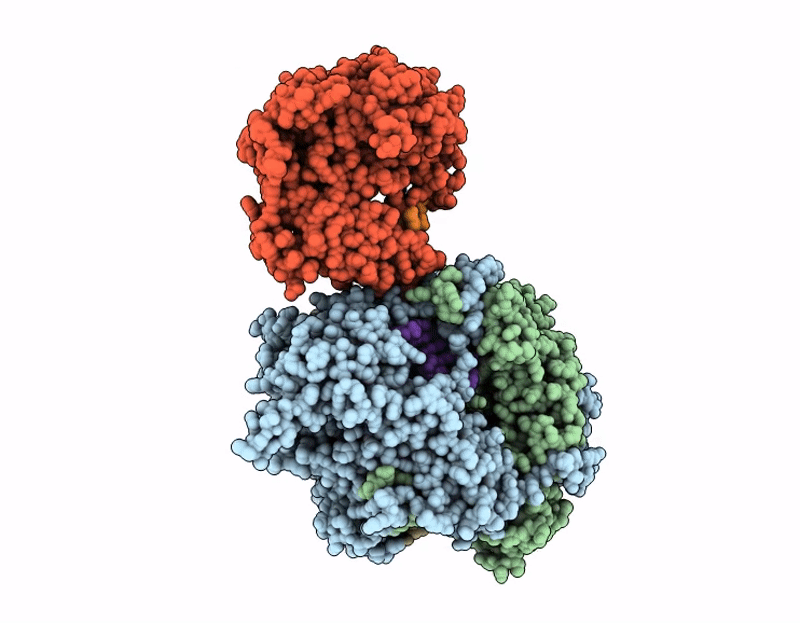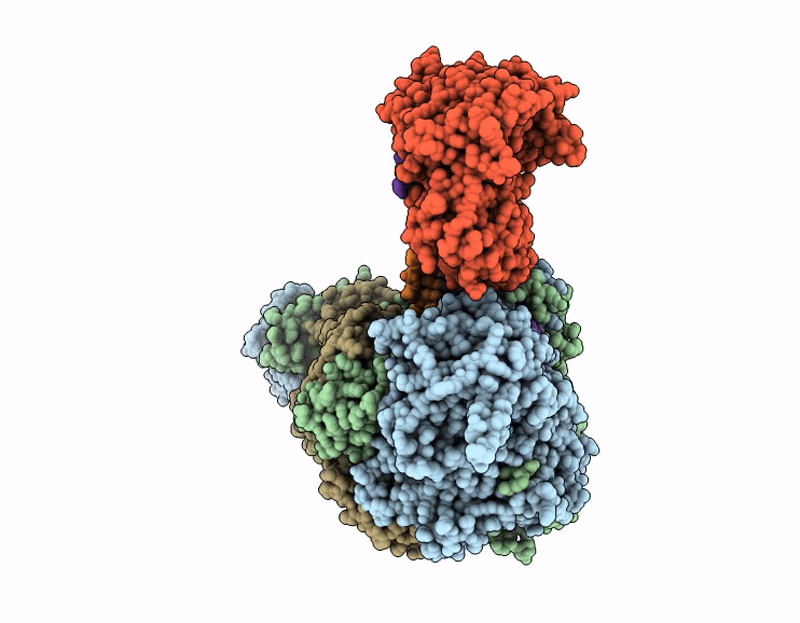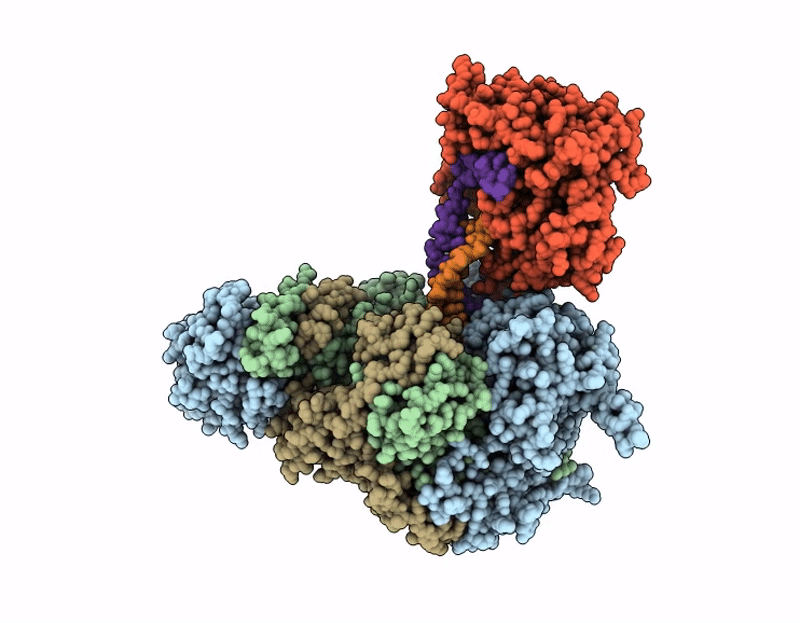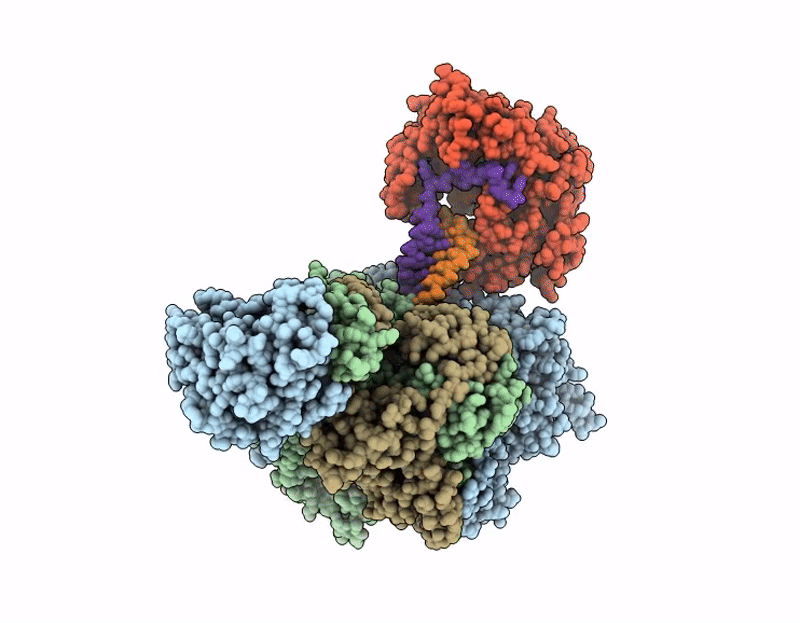
Title:
Coupling of polymerase-nucleoprotein-RNA in an influenza virus mini ribonucleoprotein complex
Biological Source:
Source Organism(s):
Influenza A virus (A/Victoria/3/1975(H3N2)) (Taxon ID: 392809)
Expression System(s):
Method Details:
Experimental Method:
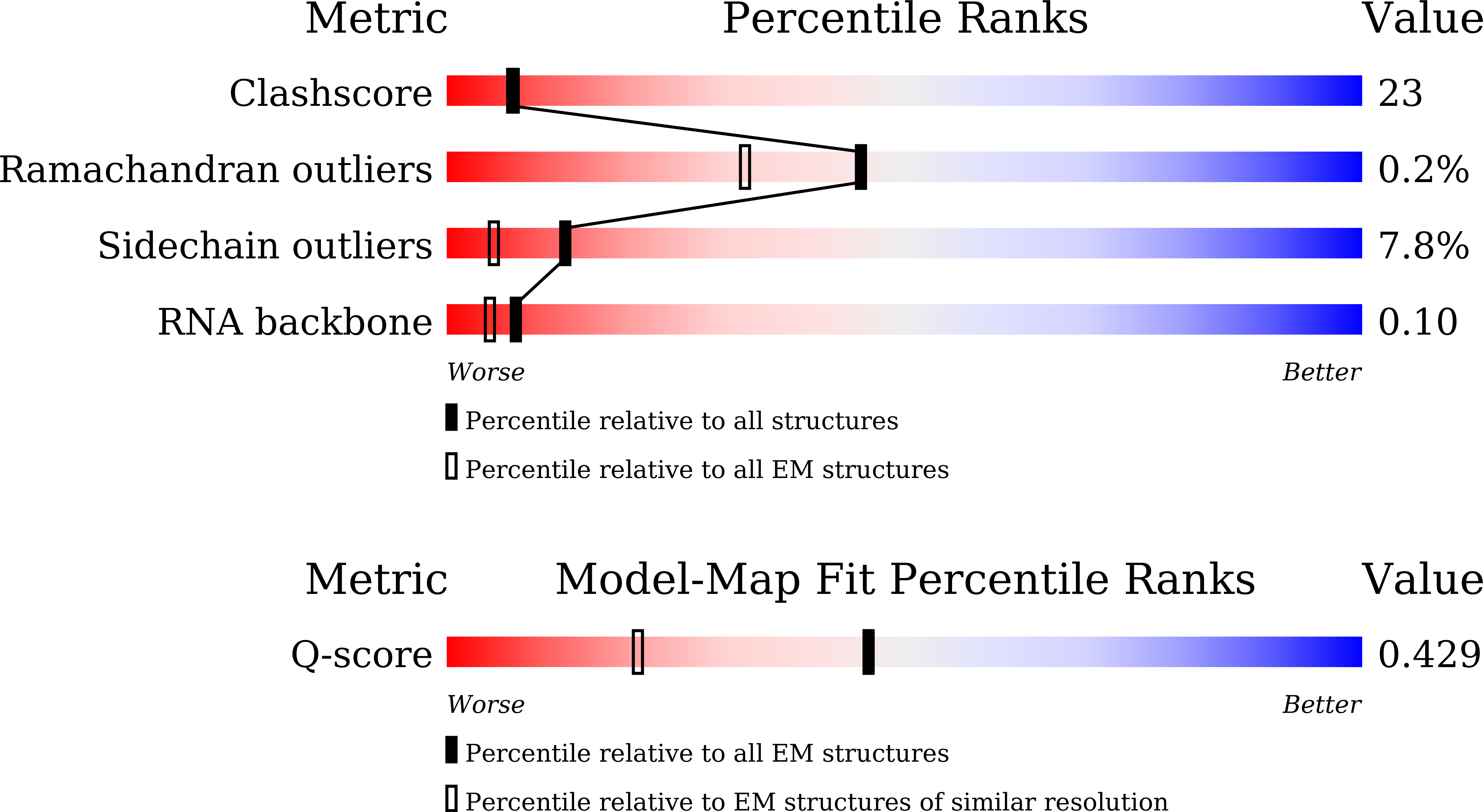
Resolution:
2.97 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE