Deposition Date
2025-04-18
Release Date
2025-08-13
Last Version Date
2025-08-13
Entry Detail
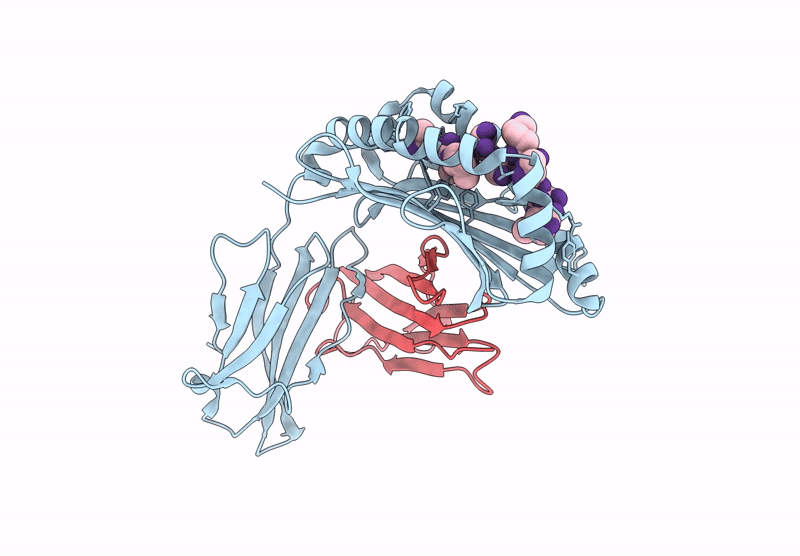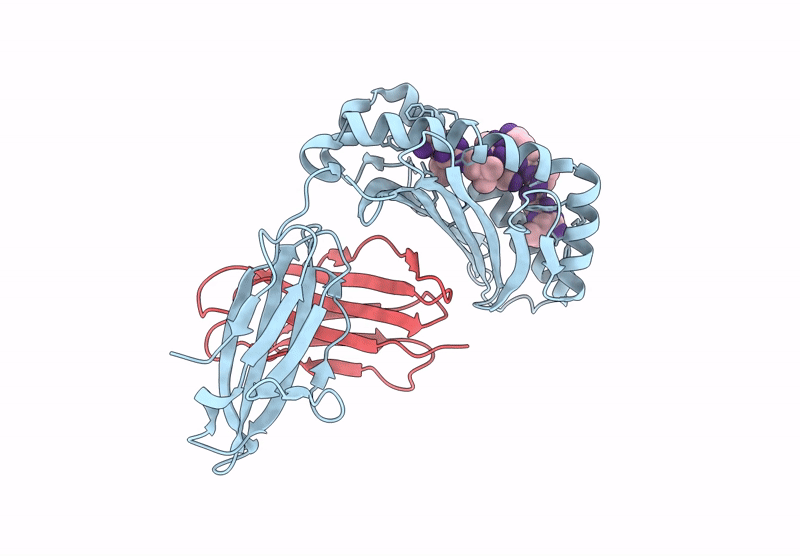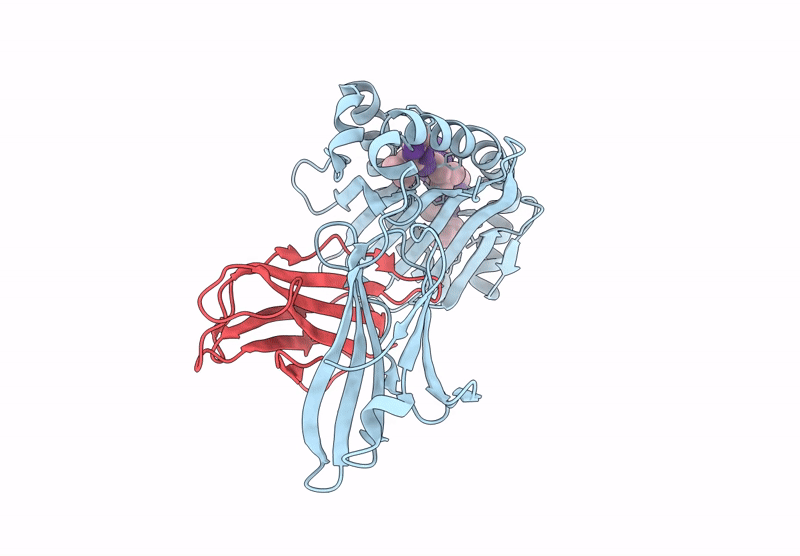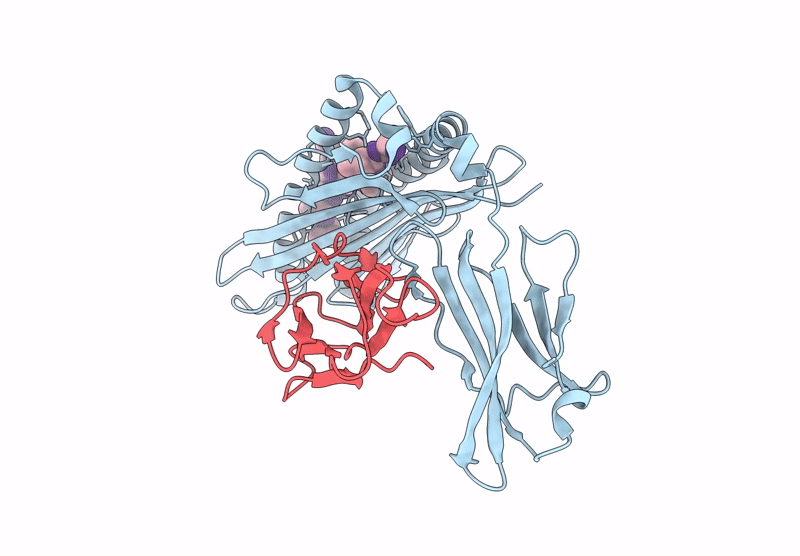
PDB ID:
9UL1
Keywords:
Title:
Crystal structure of Tibetan wild boar SLA-1*Z0301 for 2.32 angstrom
Biological Source:
Source Organism(s):
Sus scrofa (Taxon ID: 9823)
Porcine reproductive and respiratory syndrome virus (Taxon ID: 28344)
Porcine reproductive and respiratory syndrome virus (Taxon ID: 28344)
Expression System(s):
Method Details:
Experimental Method:
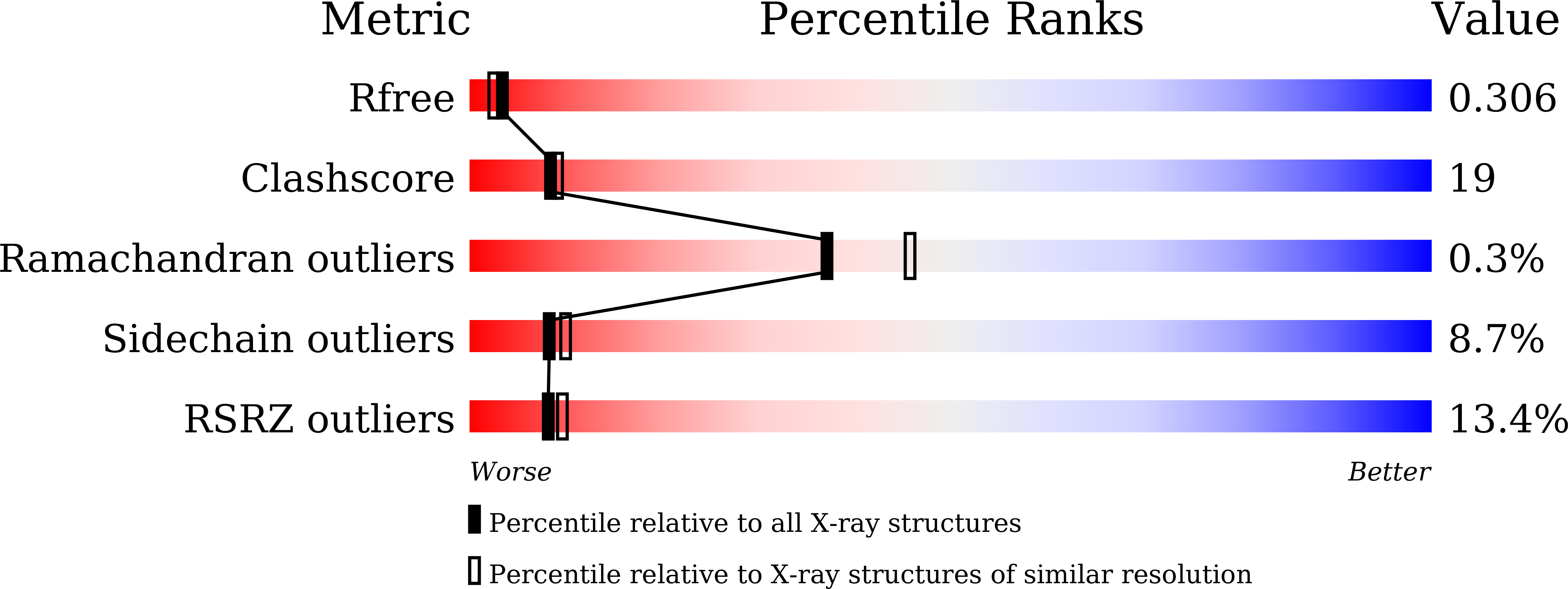
Resolution:
2.32 Å
R-Value Free:
0.29
R-Value Work:
0.27
Space Group:
C 1 2 1