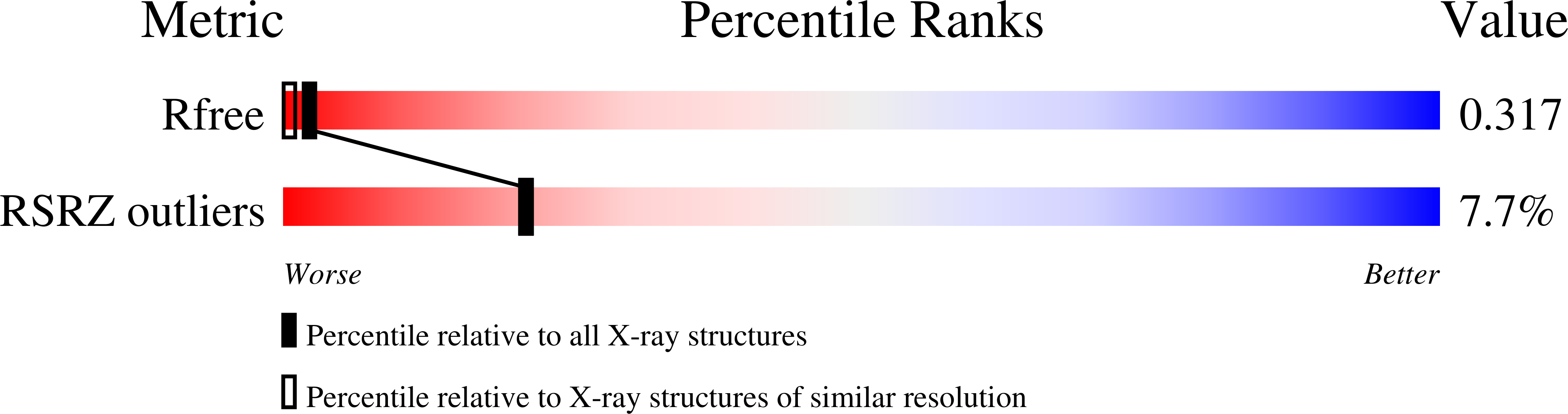
Deposition Date
2025-07-13
Release Date
2025-10-08
Last Version Date
2025-10-22
Entry Detail
PDB ID:
9RXX
Keywords:
Title:
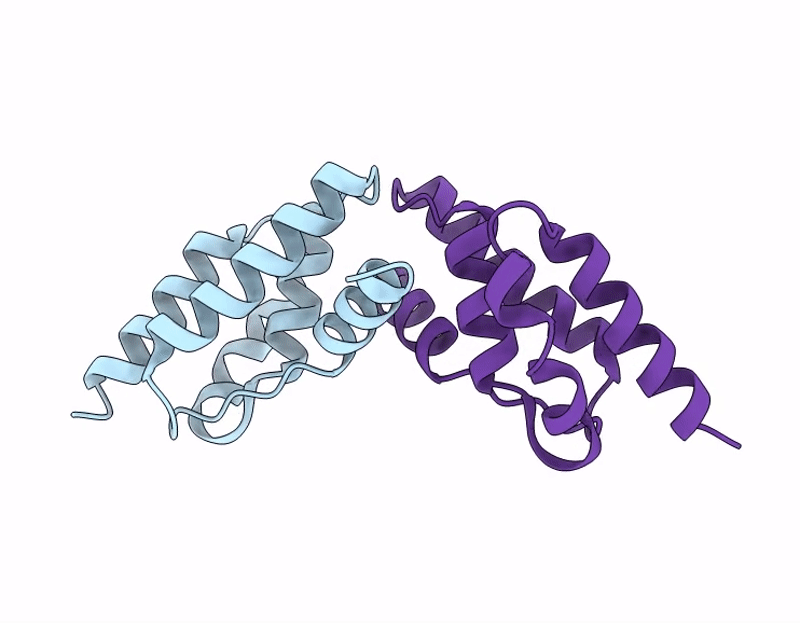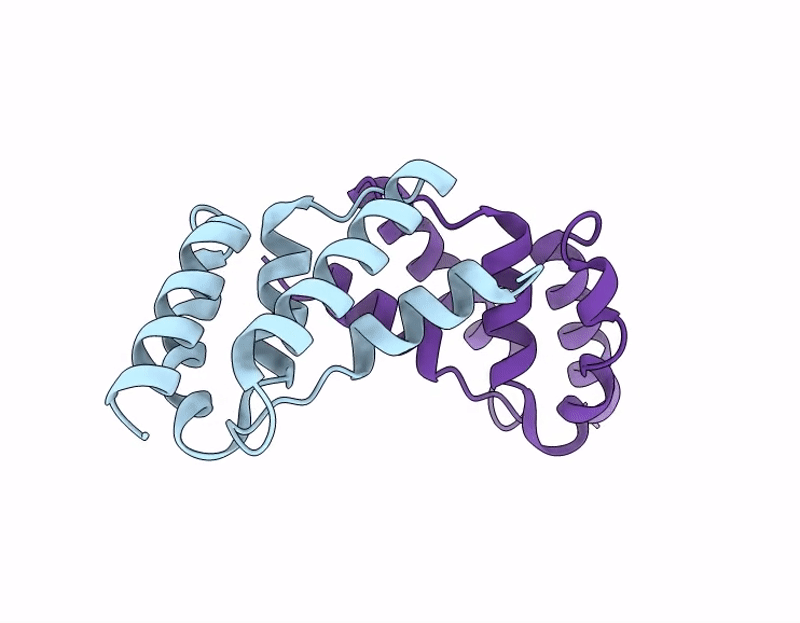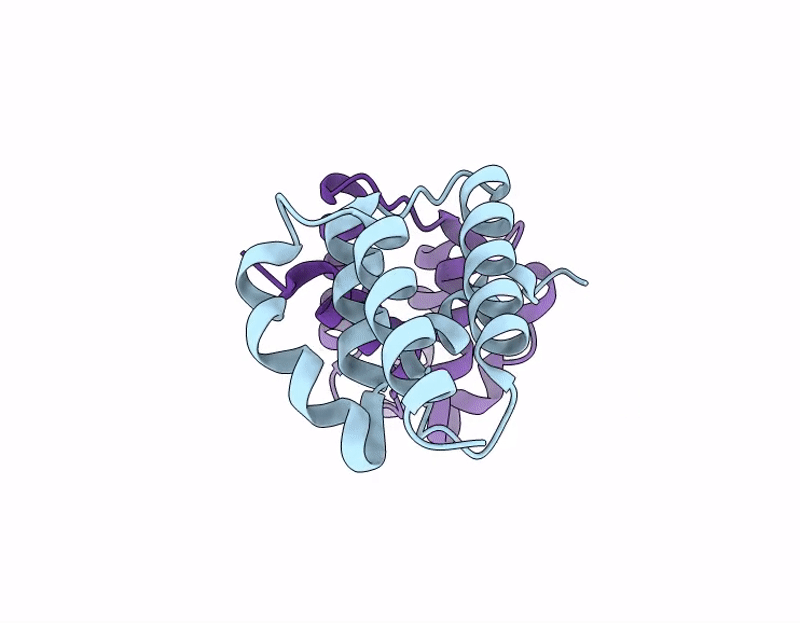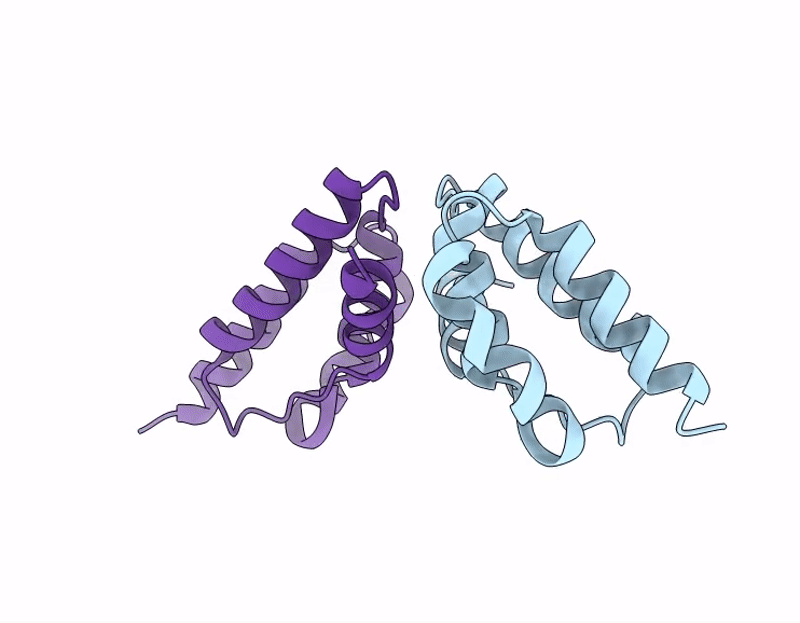
Ty1 Prime Retrotransposon Capsid C-Terminal Domain, F323S
Biological Source:
Source Organism:
Saccharomyces cerevisiae (Taxon ID: 4932)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.62 Å
R-Value Free:
0.26
R-Value Work:
0.22
Space Group:
P 1 21 1