Deposition Date
2025-07-11
Release Date
2025-11-05
Last Version Date
2025-11-05
Entry Detail
PDB ID:
9RXF
Keywords:
Title:
E20K/N28G/V36L/D43K/Q48E/I59A/E61K/E72K/V76L/N79S/I92A/D126K/A142V/D153K/D154E/S158T FLAVODOXIN FROM ANABAENA
Biological Source:
Source Organism(s):
Nostoc sp. PCC 7119 (Taxon ID: 1168)
Expression System(s):
Method Details:
Experimental Method:
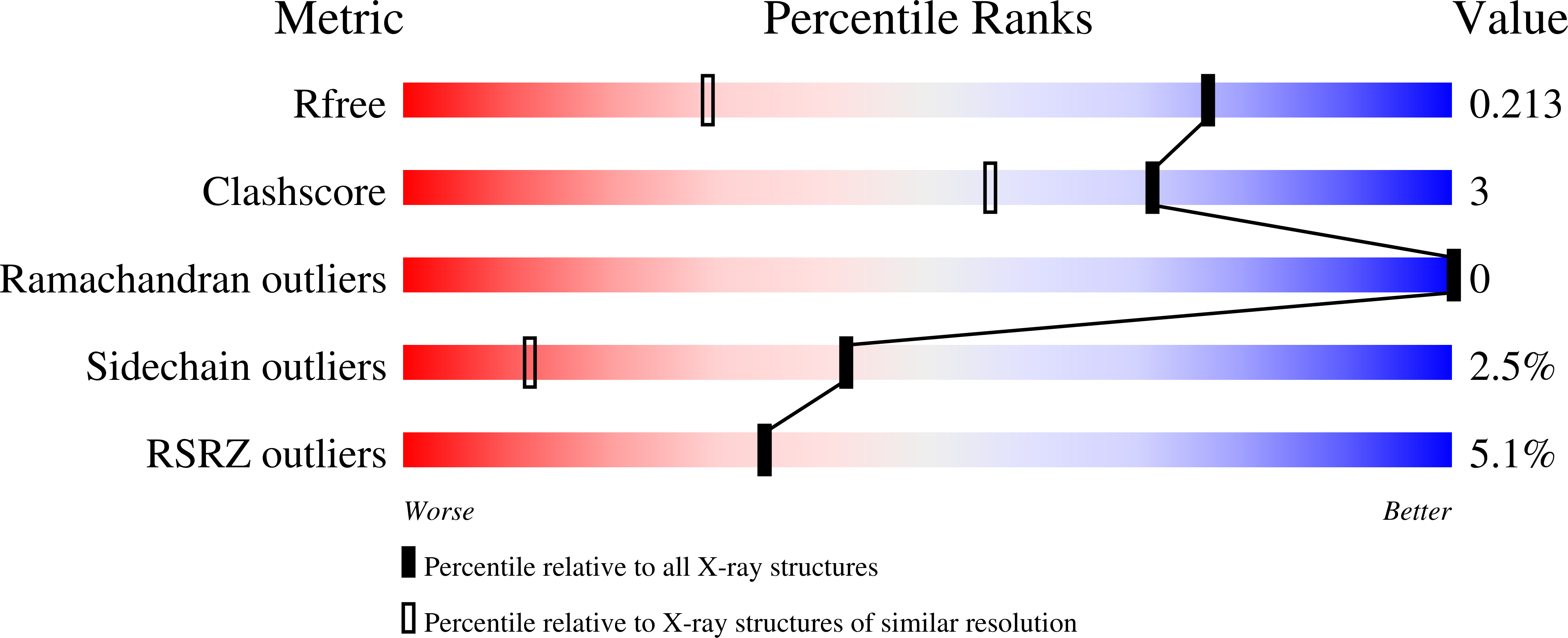
Resolution:
1.40 Å
R-Value Free:
0.20
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 21 21 21