Deposition Date
2025-06-08
Release Date
2025-09-03
Last Version Date
2025-10-22
Entry Detail
PDB ID:
9RGW
Keywords:
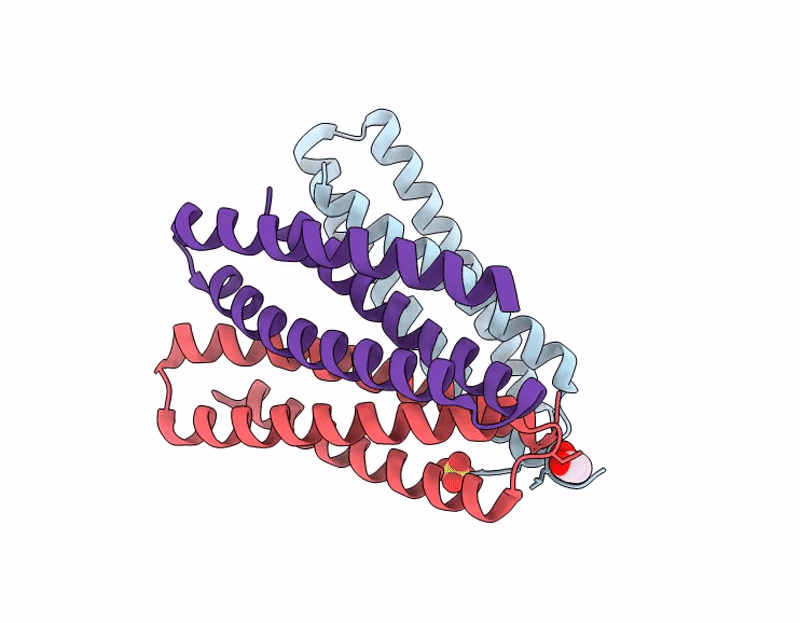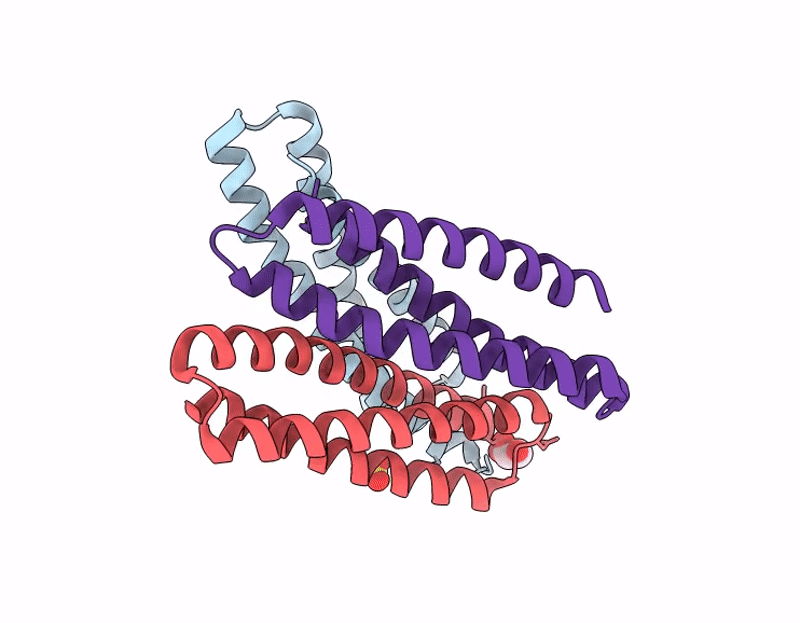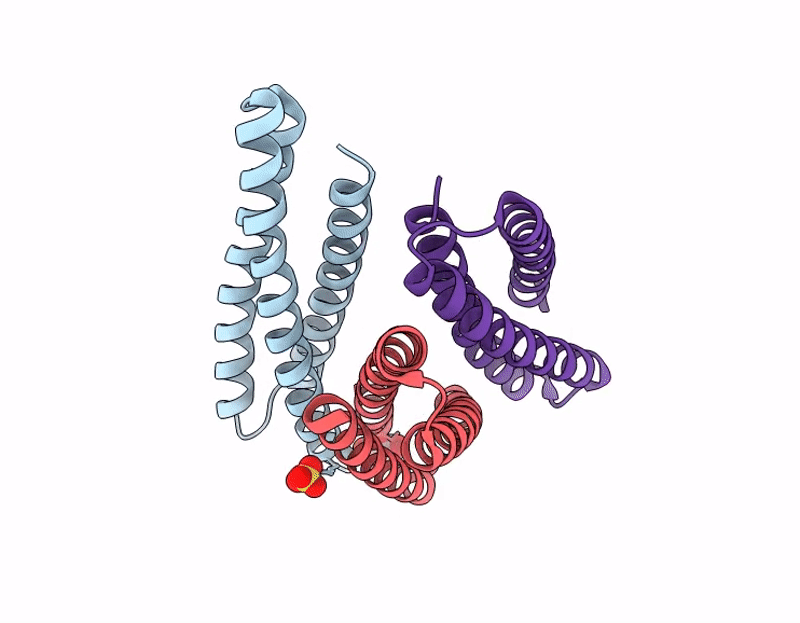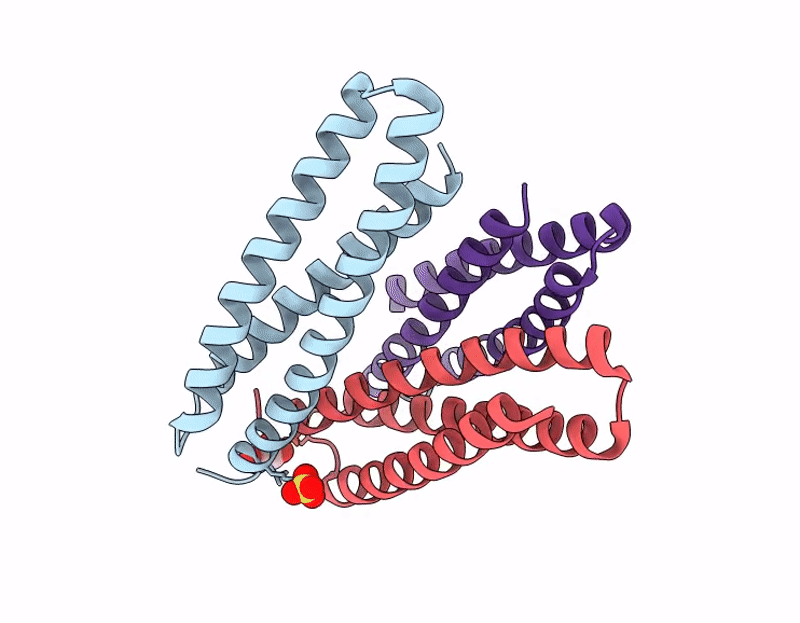
Title:
X-ray crystal structure of a de novo designed single-chain antiparallel 3-helix coiled-coil bundle, sc-apCC3-CW2
Biological Source:
Source Organism:
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
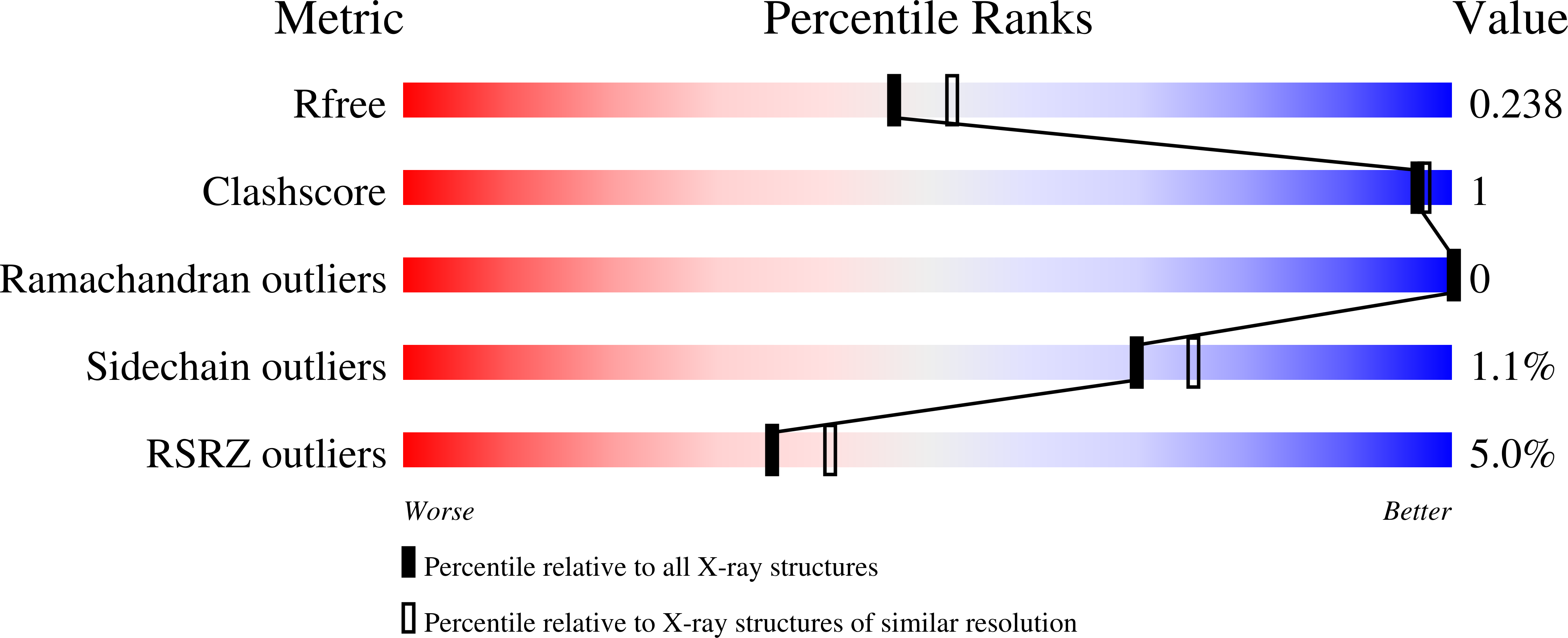
Resolution:
2.15 Å
R-Value Free:
0.23
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 41 21 2