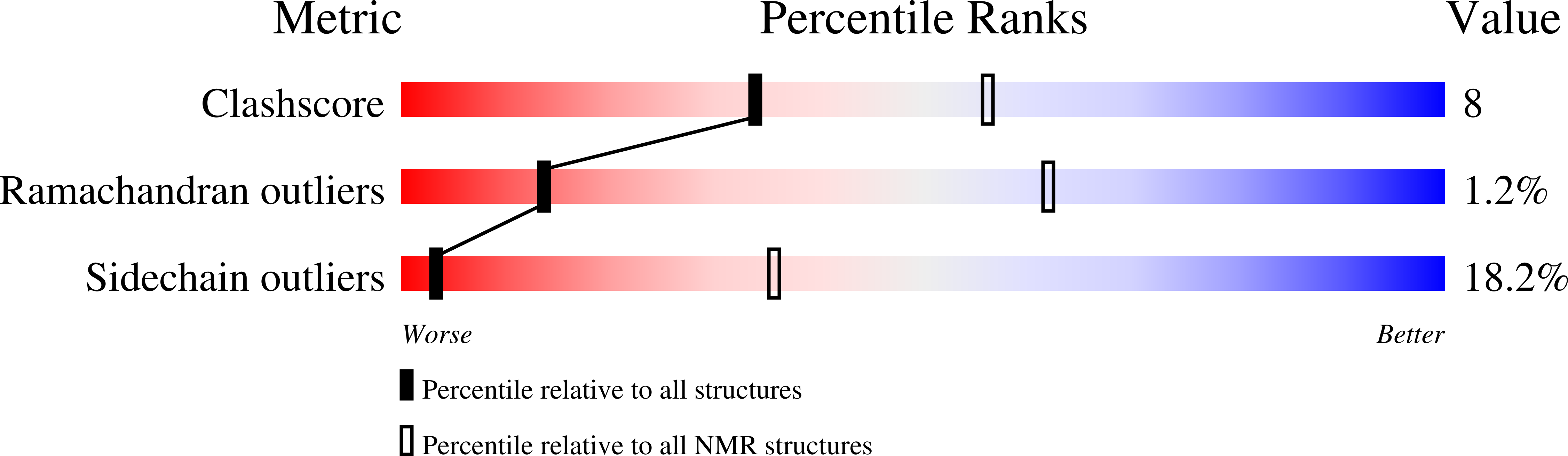
Deposition Date
2025-04-14
Release Date
2025-08-13
Last Version Date
2025-09-17
Entry Detail
PDB ID:
9QWI
Keywords:
Title:
The N-terminal domain (44-180) of the SARS-CoV-2 nucleocapsid phosphoprotein using an automatic assignment/modeling software
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
target function