Deposition Date
2025-03-13
Release Date
2025-04-02
Last Version Date
2025-05-14
Entry Detail
PDB ID:
9QGE
Keywords:
Title:
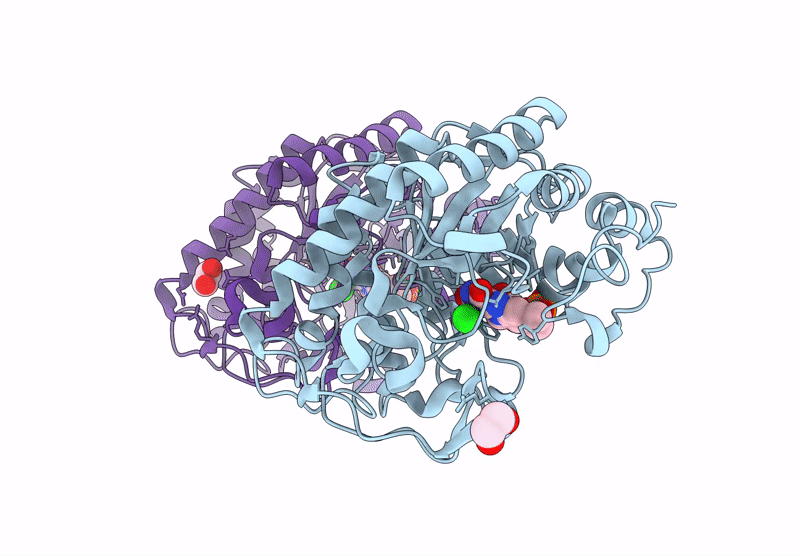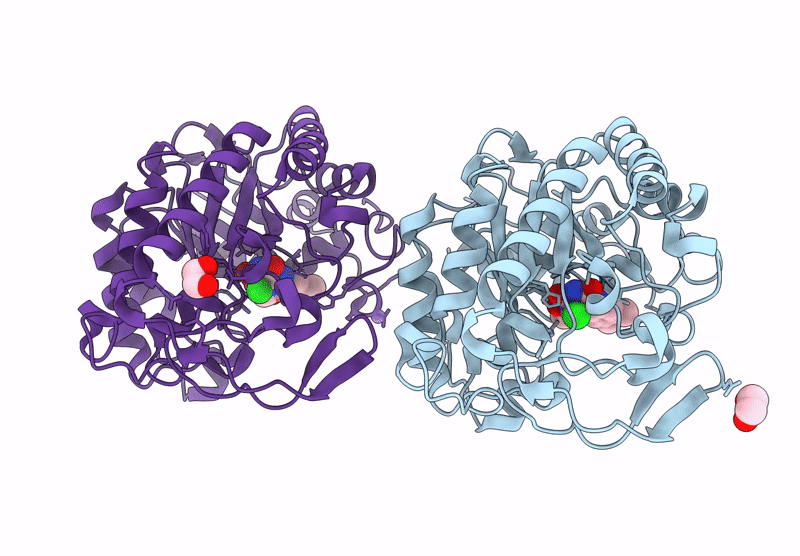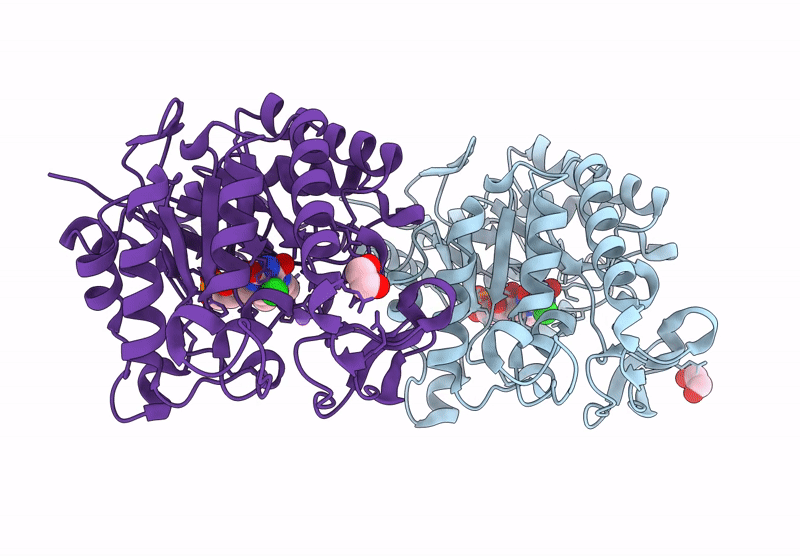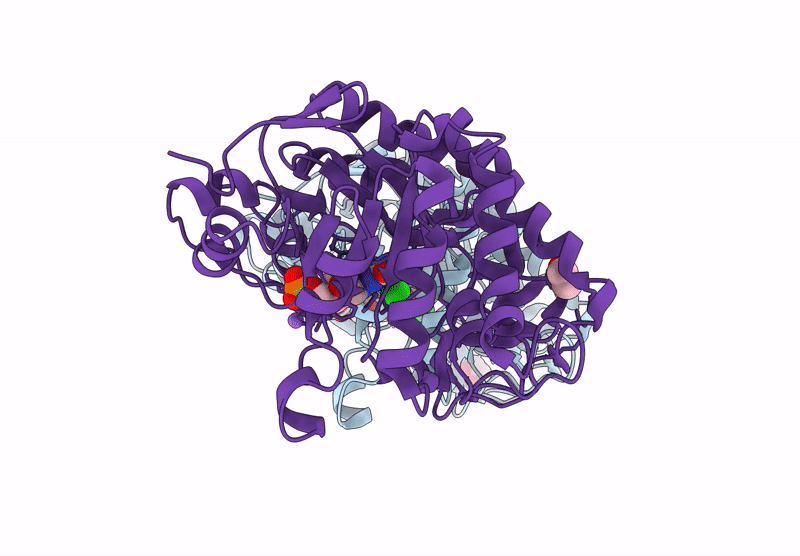
Crystal structure of an NADH-accepting ene reductase variant NostocER1-L1,5 mutant D352K
Biological Source:
Source Organism(s):
Nostoc sp. PCC 7120 = FACHB-418 (Taxon ID: 103690)
Expression System(s):
Method Details:
Experimental Method:
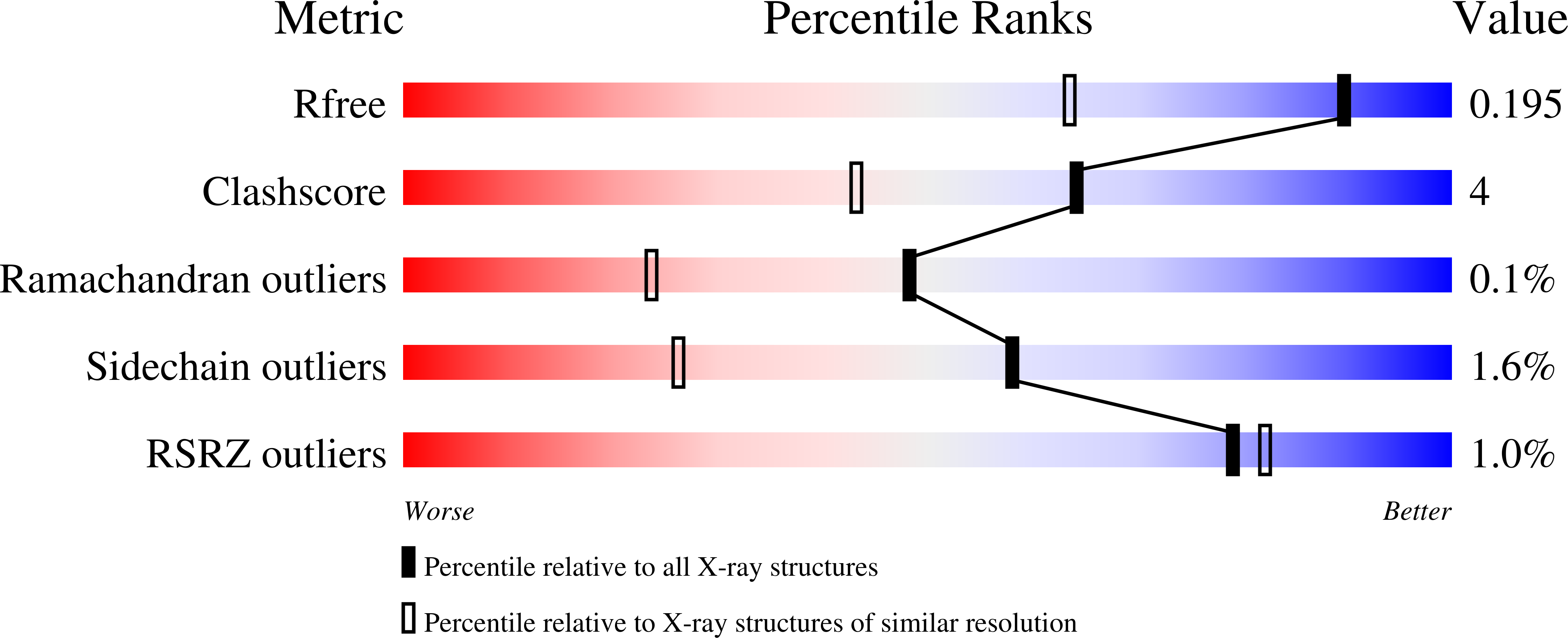
Resolution:
1.43 Å
R-Value Free:
0.19
R-Value Work:
0.15
Space Group:
P 1 21 1