Deposition Date
2025-02-23
Release Date
2025-05-14
Last Version Date
2025-11-19
Entry Detail
PDB ID:
9Q87
Keywords:
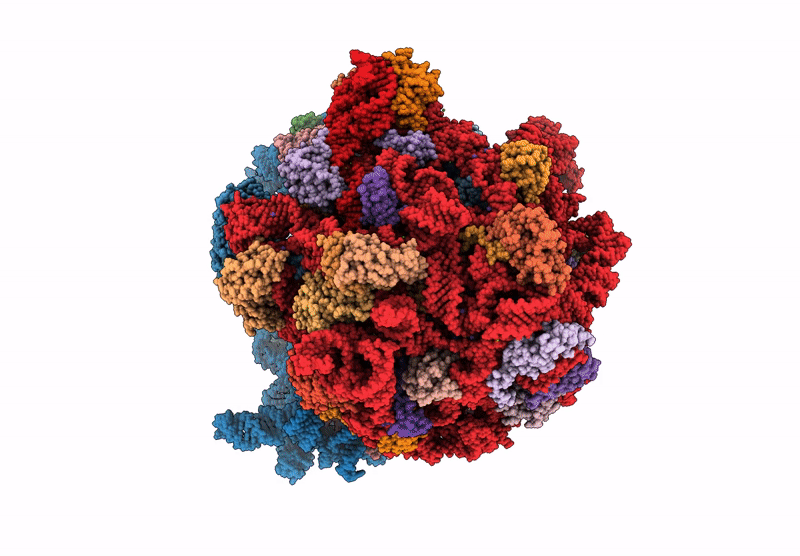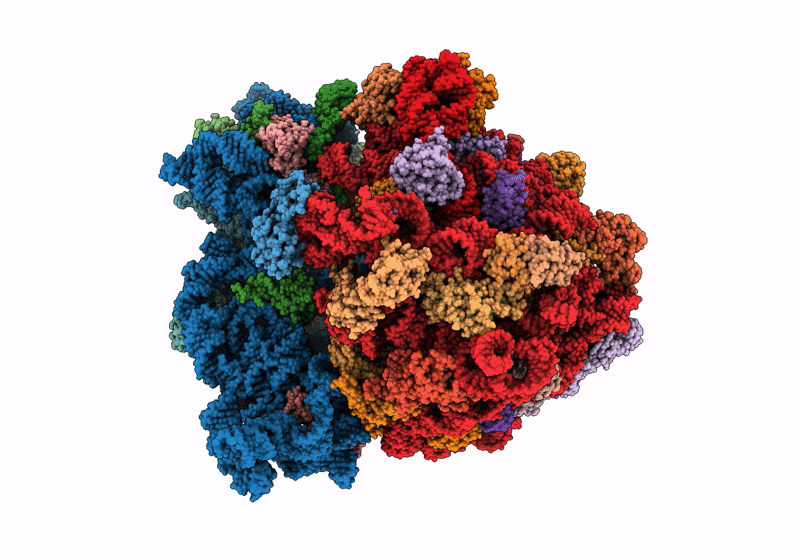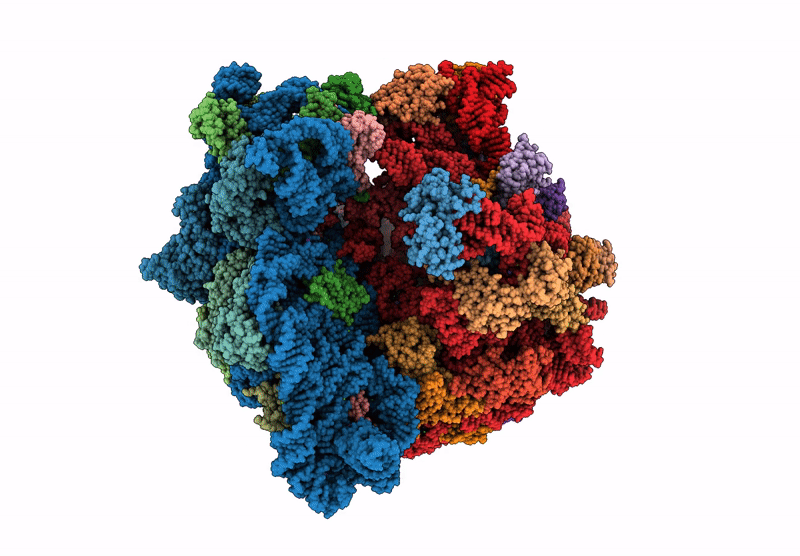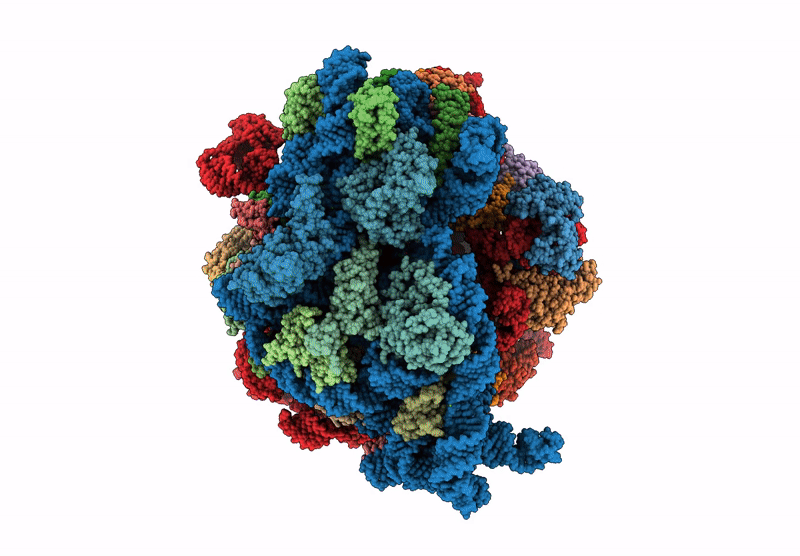
Title:
Principles of ion binding to RNA inferred from the analysis of a 1.55 Angstrom resolution bacterial ribosome structure - Part I: Mg2+
Biological Source:
Source Organism(s):
Escherichia coli B (Taxon ID: 37762)
Method Details:
Experimental Method:
Resolution:
1.55 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


