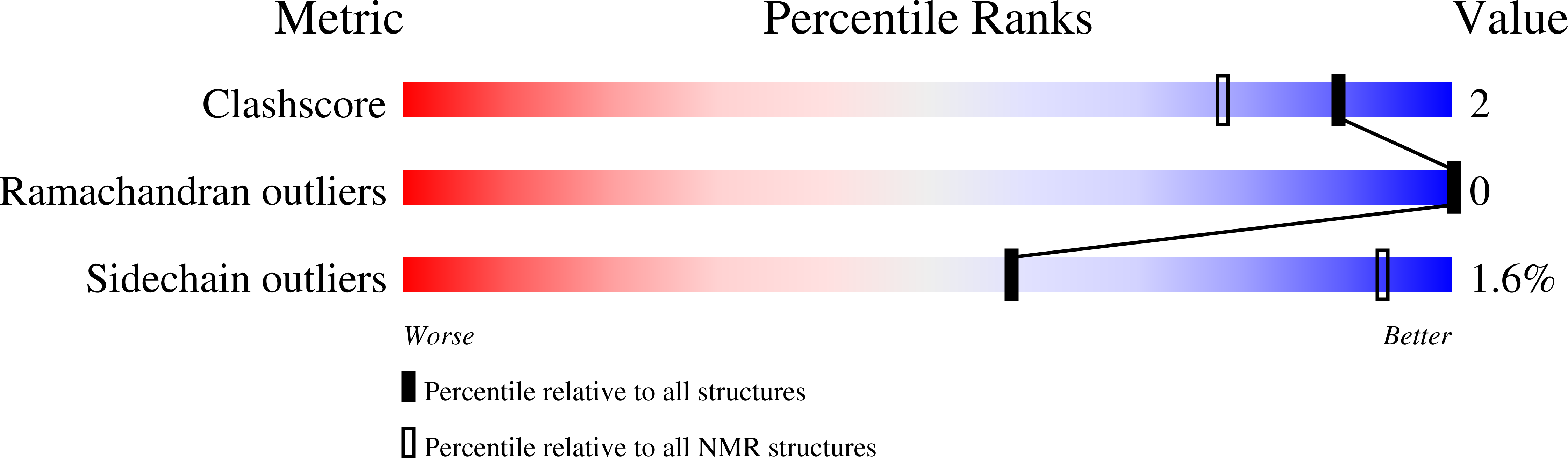
Deposition Date
2025-08-08
Release Date
2025-09-17
Last Version Date
2026-02-18
Entry Detail
PDB ID:
9PYS
Keywords:
Title:
NMR RDC refinement of the helical domain of the SARS-CoV-2 monomeric Main Protease (MPROH41Q,10-306)
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
10
Conformers Submitted:
10
Selection Criteria:
all calculated structures submitted