Deposition Date
2025-07-18
Release Date
2026-01-21
Last Version Date
2026-01-21
Entry Detail
PDB ID:
9PN3
Keywords:
Title:
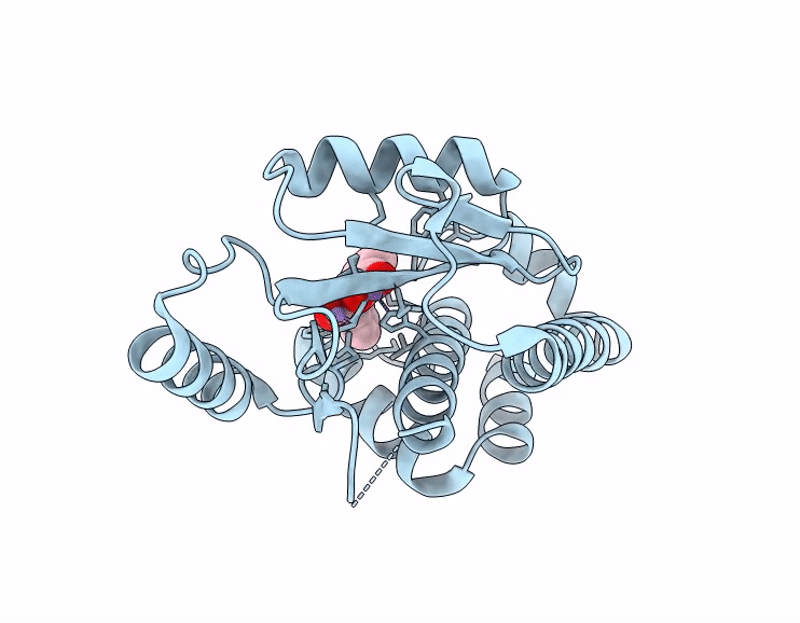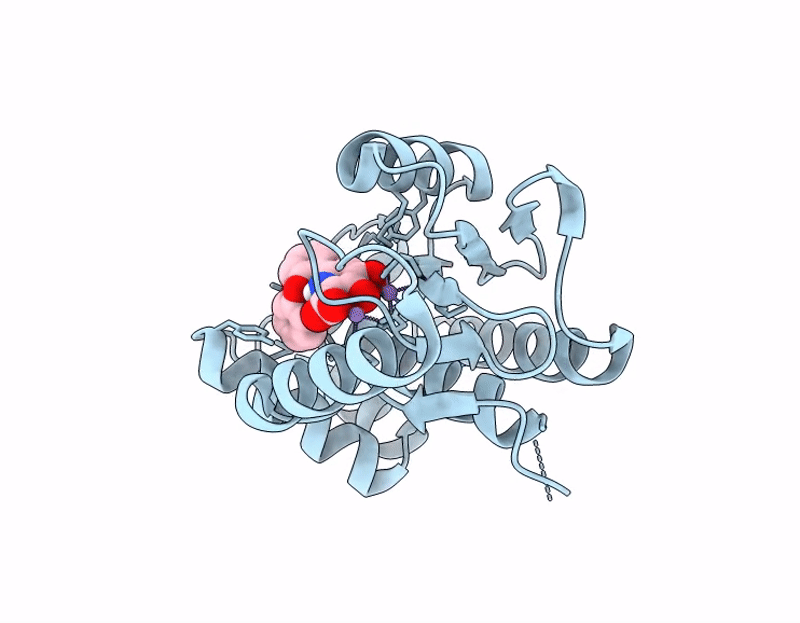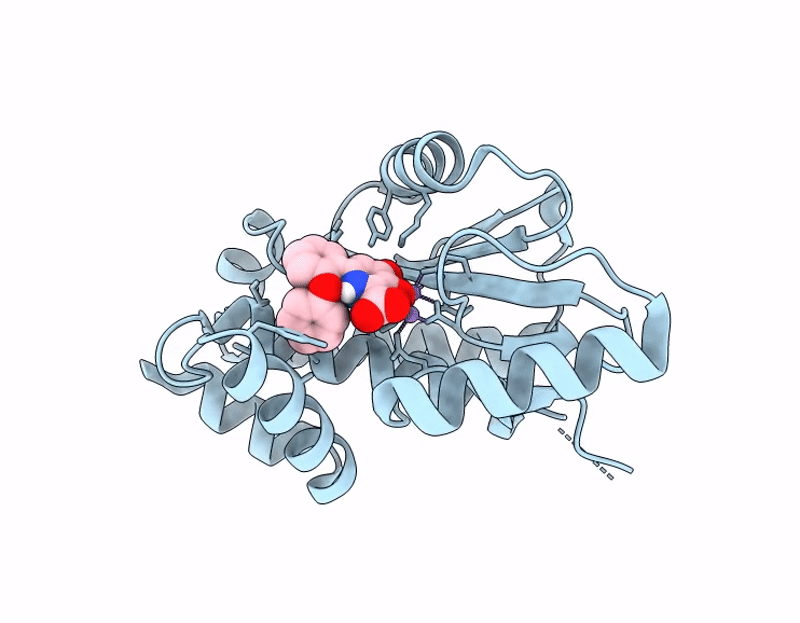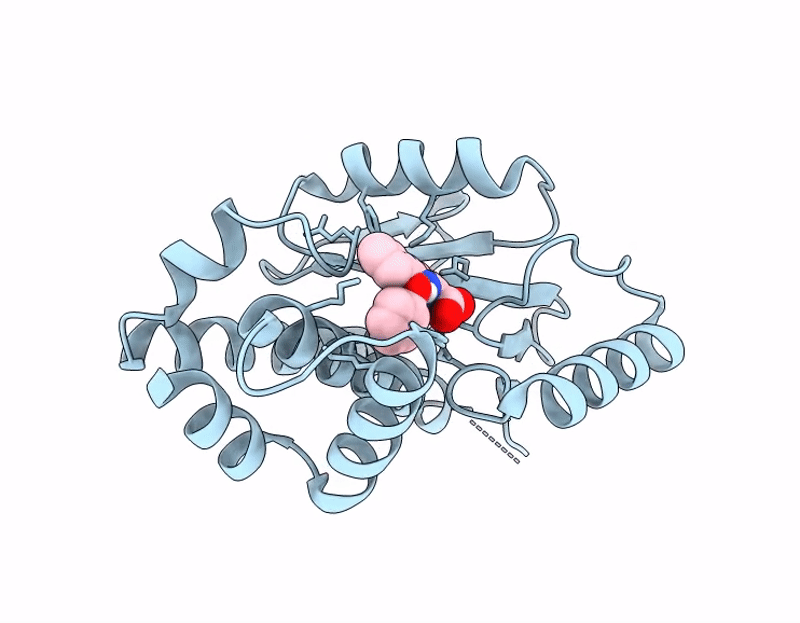
Influenza PA-N Endonuclease I38T mutant with compound 4 ((6M)-3-hydroxy-4-oxo-6-(2-phenoxyphenyl)-1,4-dihydropyridine-2-carboxylic acid)
Biological Source:
Source Organism(s):
Influenza A virus (A/California/04/2009(H1N1)) (Taxon ID: 641501)
Expression System(s):
Method Details:
Experimental Method:
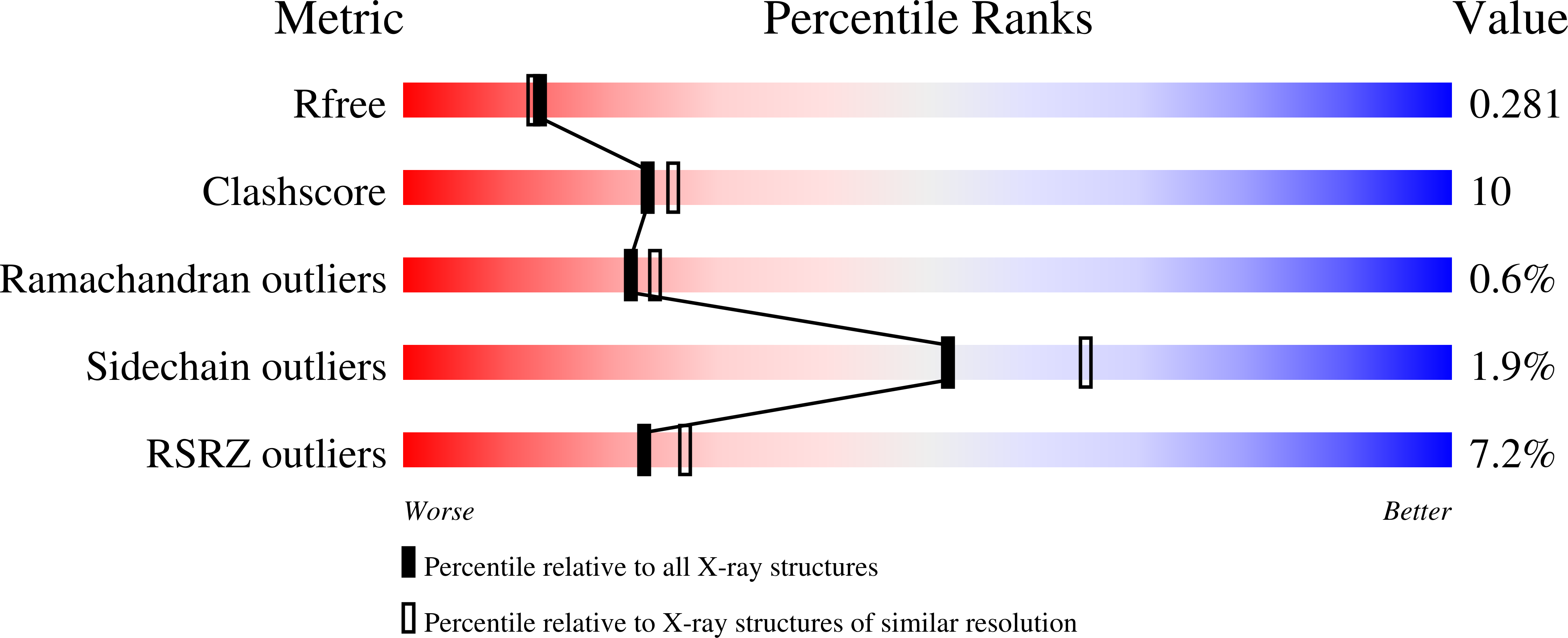
Resolution:
2.36 Å
R-Value Free:
0.27
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 62 2 2