Deposition Date
2025-07-09
Release Date
2025-10-22
Last Version Date
2025-11-26
Entry Detail
PDB ID:
9PHW
Keywords:
Title:
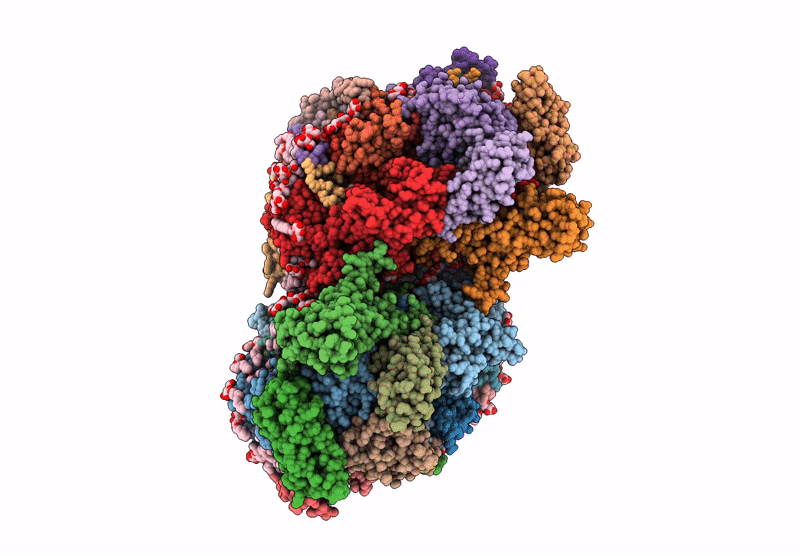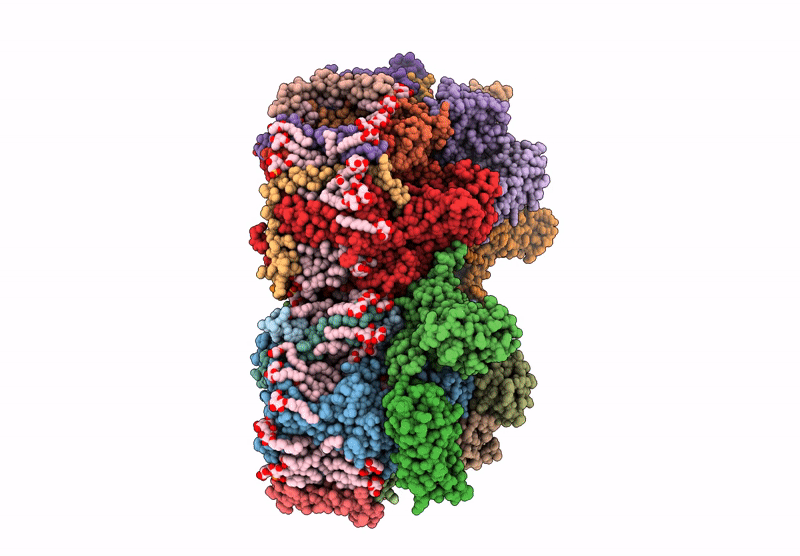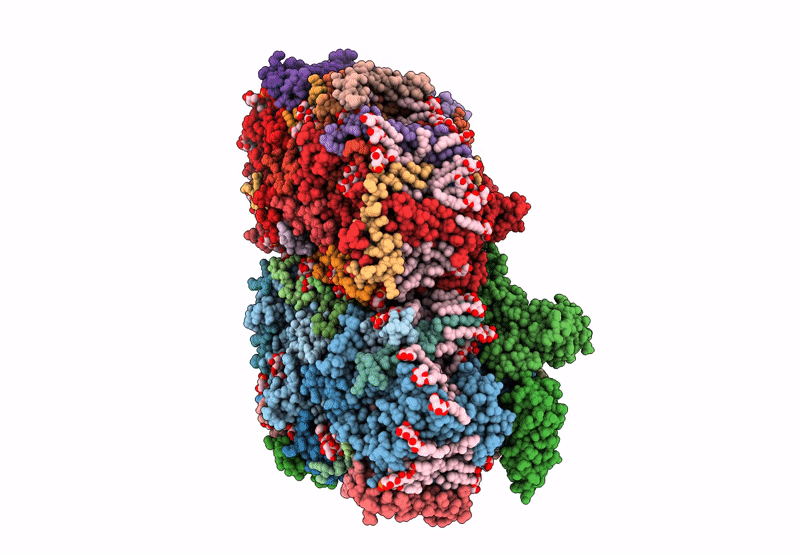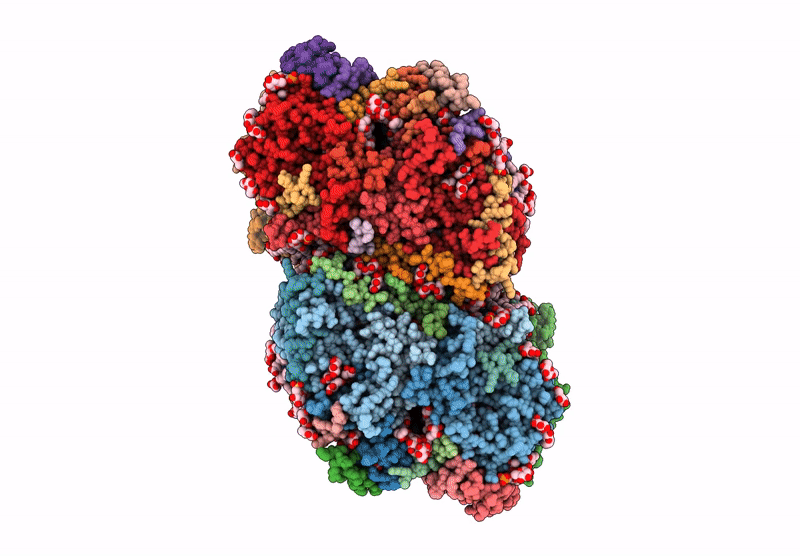
Structure of the D1-Val185Asn mutated photosystem II complex with slow O-O bond formation reveals changes in the Cl1 water channel
Biological Source:
Source Organism(s):
Synechocystis sp. PCC 6803 (Taxon ID: 1148)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.99 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


