Deposition Date
2025-06-30
Release Date
2025-11-19
Last Version Date
2025-12-24
Entry Detail
PDB ID:
9PDI
Keywords:
Title:
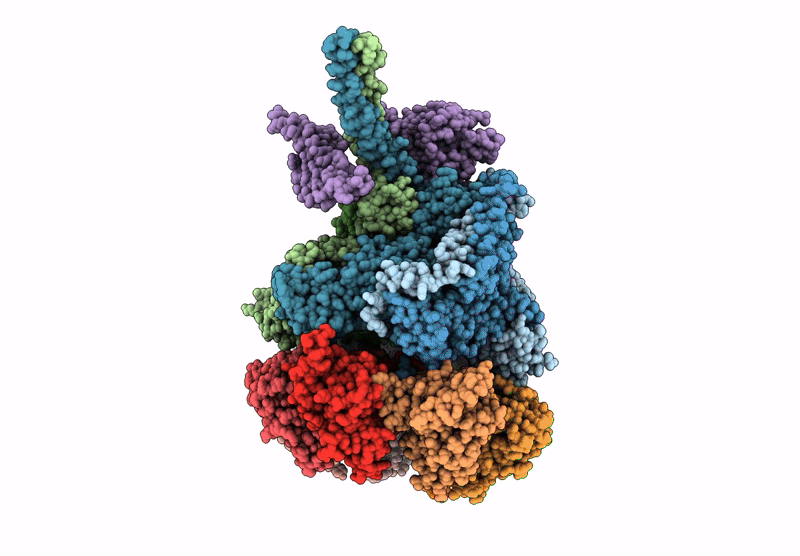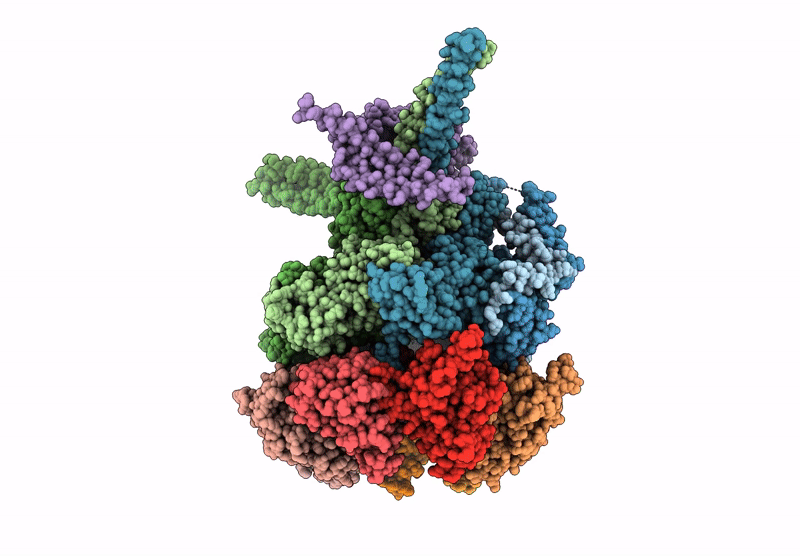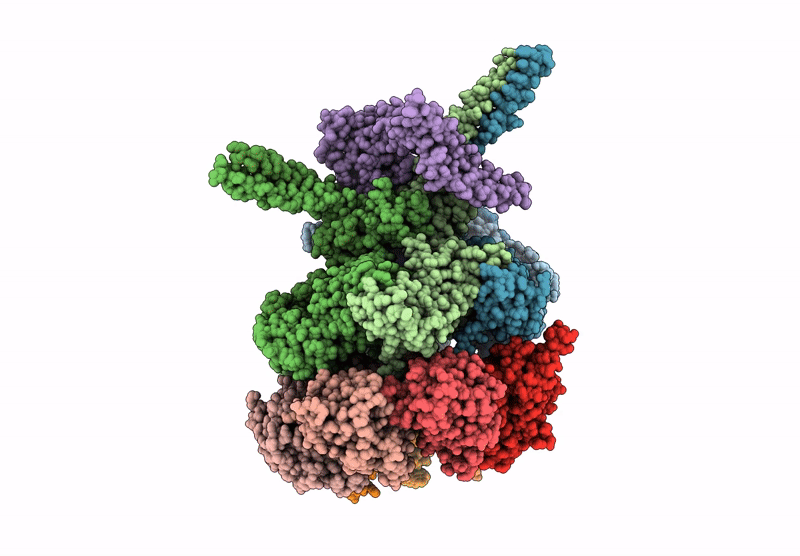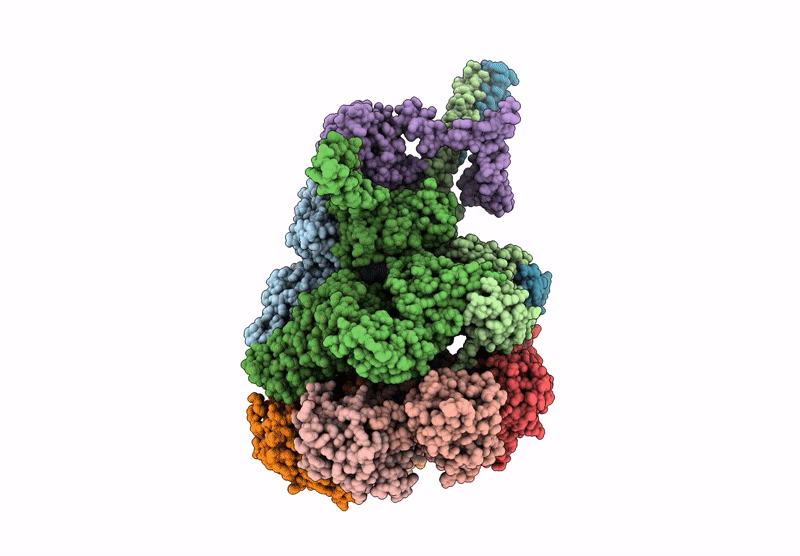
Nub1/Fat10-processing human 26S proteasome with Rpt2 at top of spiral staircase and partially unfolded Eos (AAA+ motor locally refined)
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
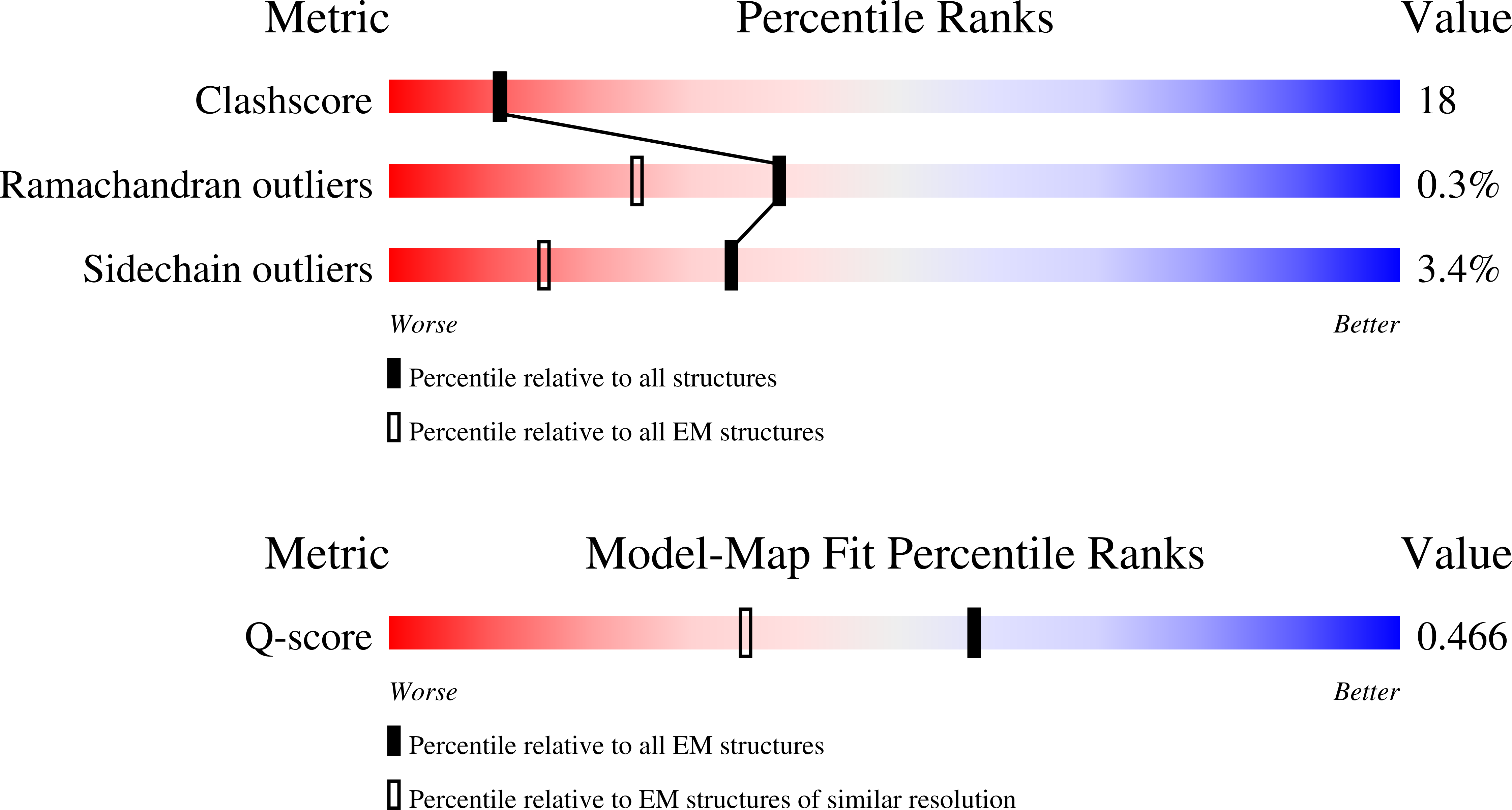
Resolution:
2.98 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE