Deposition Date
2025-06-10
Release Date
2025-11-19
Last Version Date
2025-12-10
Entry Detail
PDB ID:
9P1I
Keywords:
Title:
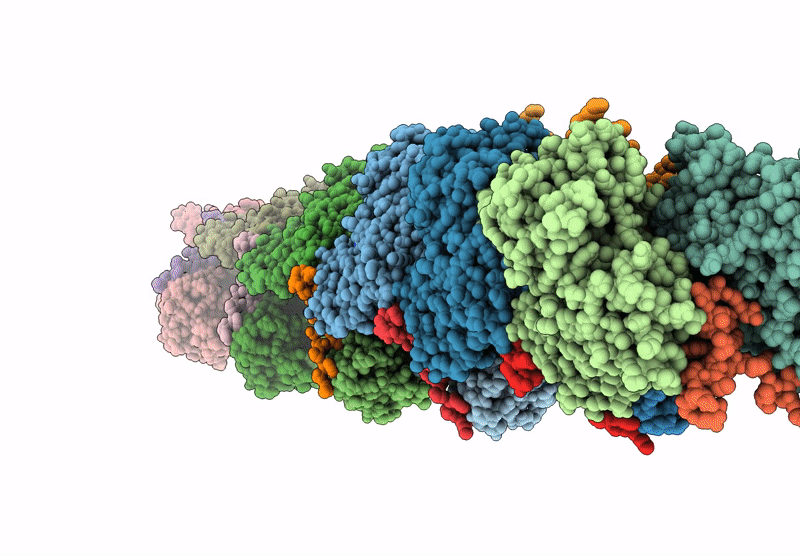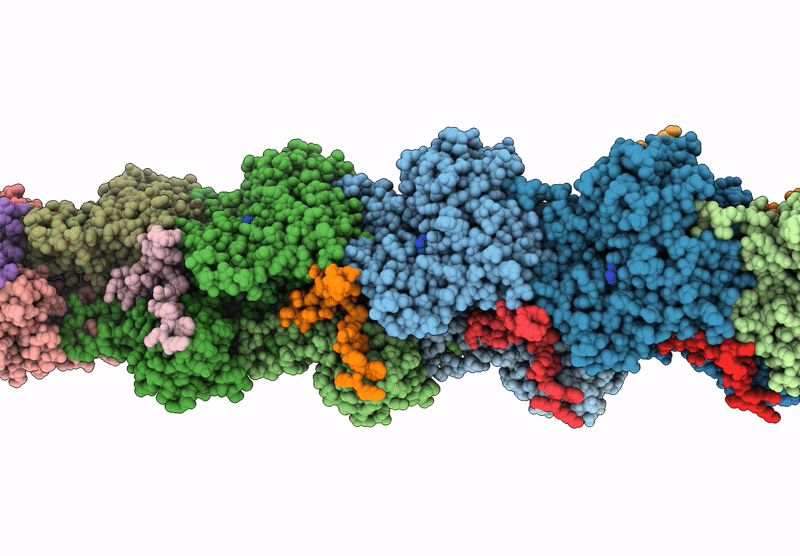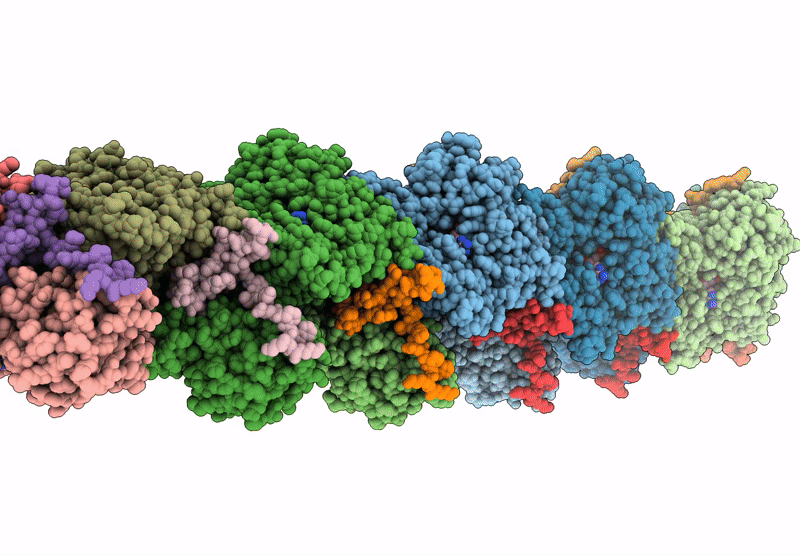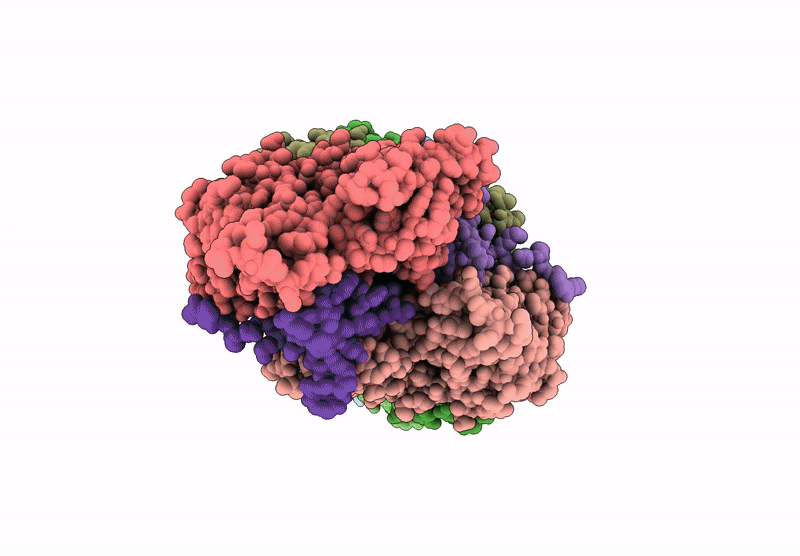
Atomic structure of vibrio effector fragment VopV bound to Beta-cytoplasmic/gamma1-cytoplasmic F-actin
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Vibrio parahaemolyticus (Taxon ID: 670)
Vibrio parahaemolyticus (Taxon ID: 670)
Expression System(s):
Method Details:
Experimental Method:
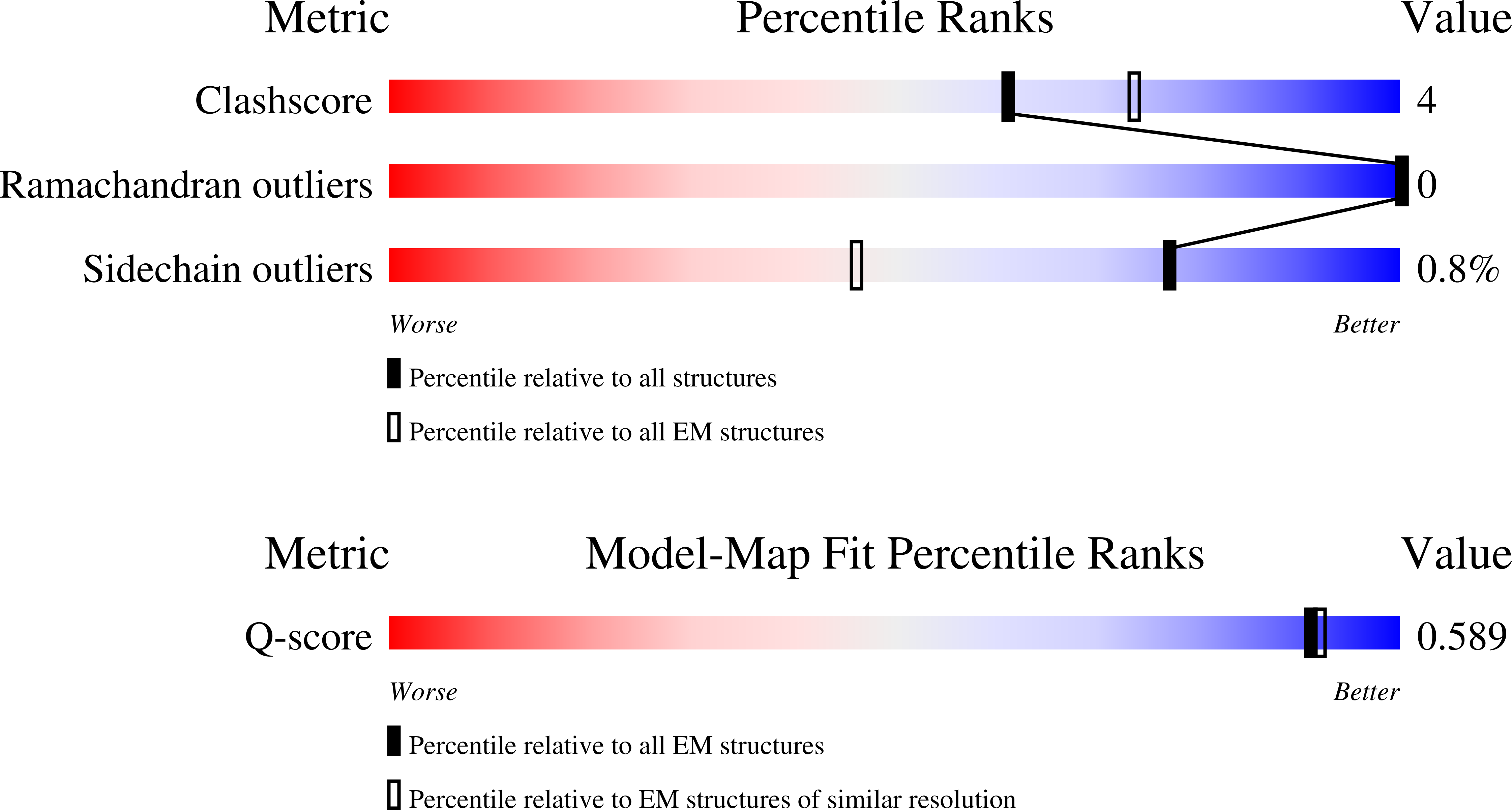
Resolution:
3.00 Å
Aggregation State:
FILAMENT
Reconstruction Method:
HELICAL