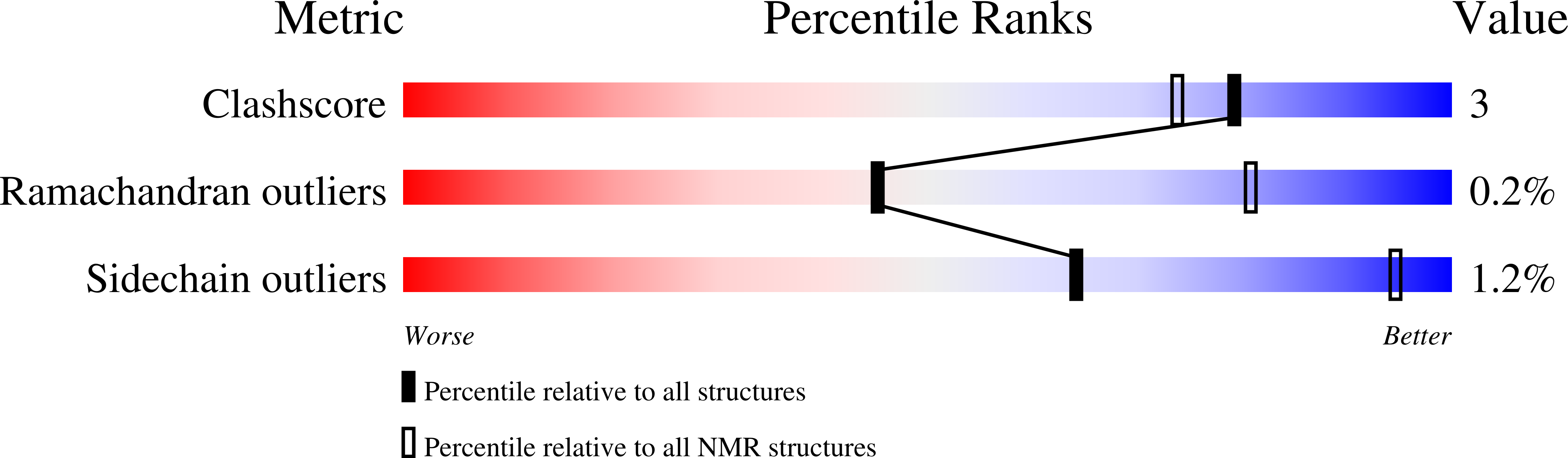
Deposition Date
2025-05-06
Release Date
2025-08-20
Last Version Date
2025-09-10
Entry Detail
PDB ID:
9OIU
Keywords:
Title:
Soltion Structure of His6-Small Ubiquitin-like Modifier (His6-SUMO)
Biological Source:
Source Organism:
Lactococcus garvieae (Taxon ID: 1363)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
1000
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy