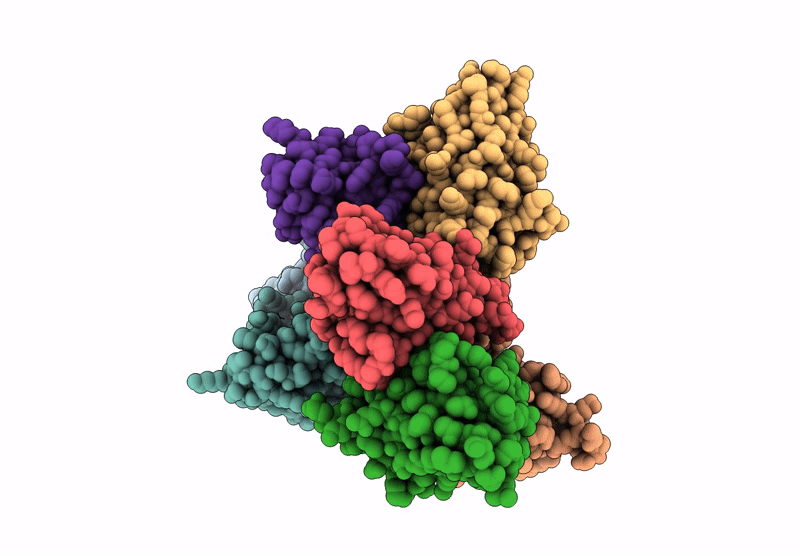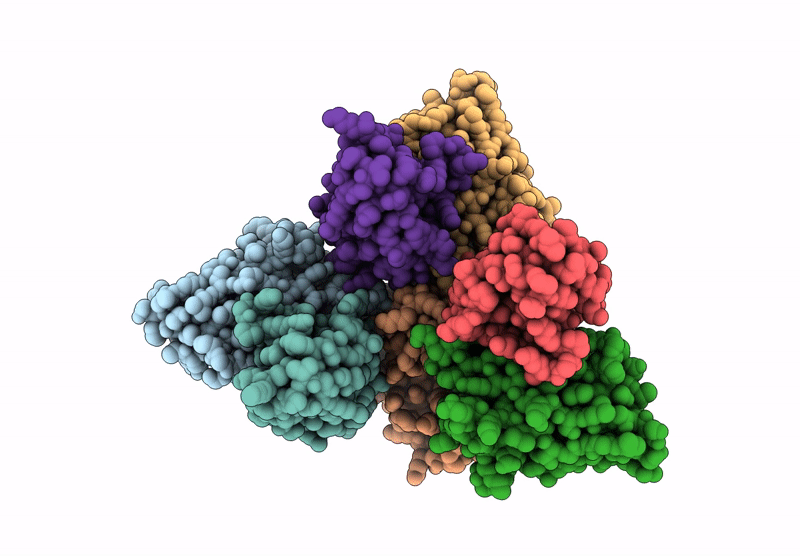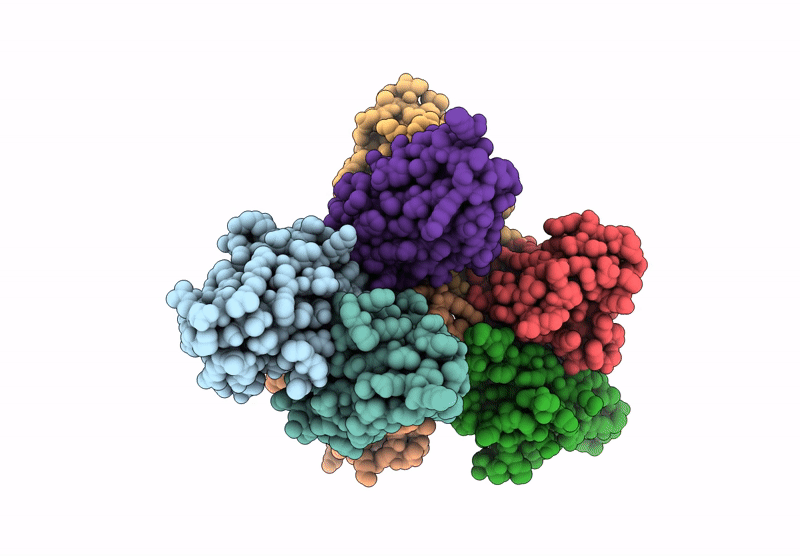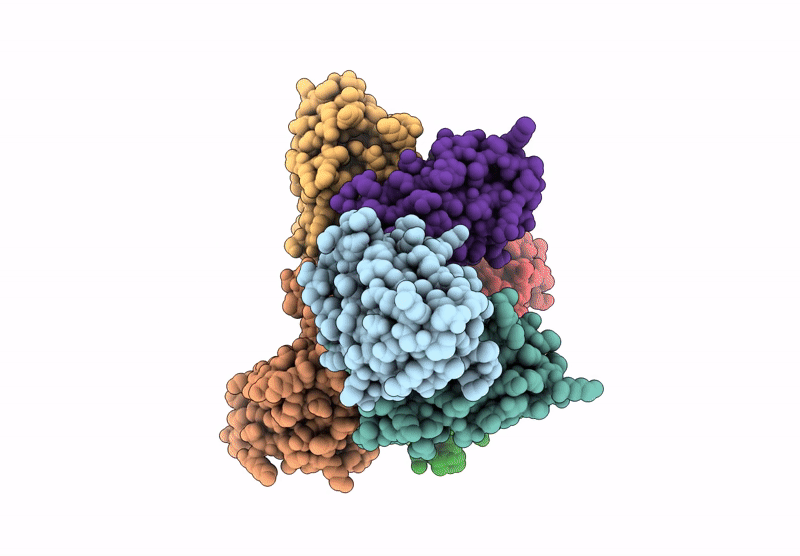
Abstact
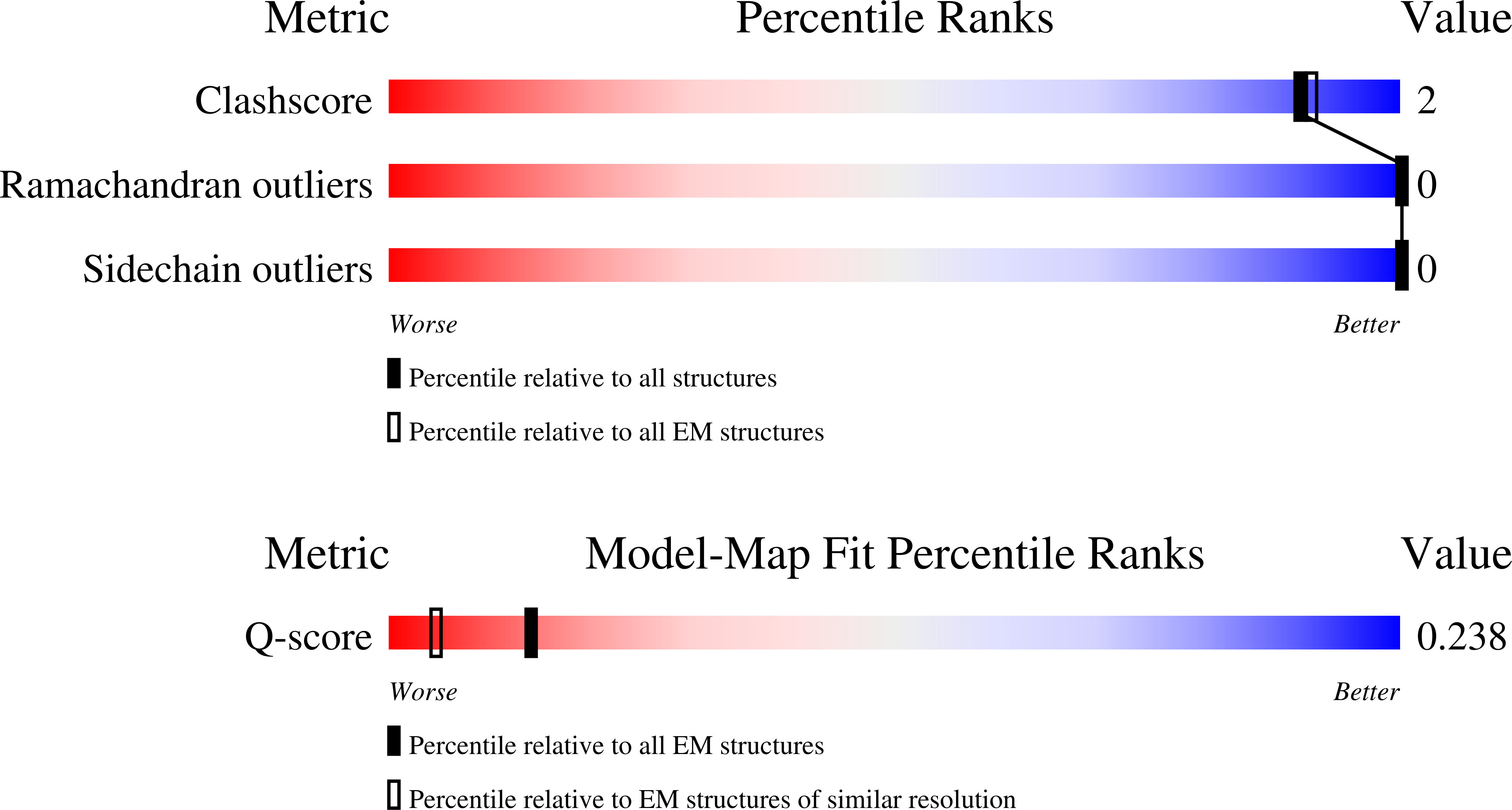
Currently approved vaccines for the prevention of malaria provide only partial protection against disease due to high variability in the quality of induced antibodies. These vaccines present the unstructured central repeat region, as well as the C-terminal domain, of the circumsporozoite protein (PfCSP) of the malaria parasite, Plasmodium falciparum [K. L. Williams et al., Nat. Med. 30,1-13 (2024)]. A recently discovered protective monoclonal antibody, L9, recognizes three structured copies of the PfCSP minor repeat. Similarly to other highly potent antimalarial antibodies, L9 relies on critical homotypic interactions between antibodies for its high protective efficacy [P. Tripathi et al., Structure 31, 480-491.e4 (2023); G. M. Martin et al., Nat. Commun. 14,2815 (2023)]. Here, we report the design of antigens scaffolding one copy of PfCSP's minor repeat capable of binding L9. To design antigens capable of presenting multiple, structure-based epitopes in one scaffold, we developed a machine learning- driven structural antigen design pipeline, MESODID, tailored to focus on multiepitope vaccine targets. We use this pipeline to design multiple scaffolds that present three copies of the PfCSP minor repeat. A 3.6 Å cryo-EM structure of our top design, minor repeat targeting immunogen (M-TIM), demonstrates that M-TIM successfully orients three copies of L9, effectively recapitulating its critical homotypic interactions. The wide prevalence of repeated epitopes in key vaccine targets, such as HIV-1 Envelope, SARS-CoV-2 spike, and Influenza Hemagglutinin, suggests that MESODID will have broad utility in creating antigens that incorporate such epitopes, offering a powerful approach to developing vaccines against a range of challenging infections, including malaria.