Deposition Date
2025-02-04
Release Date
2025-11-12
Last Version Date
2025-12-03
Entry Detail
PDB ID:
9N5Y
Keywords:
Title:
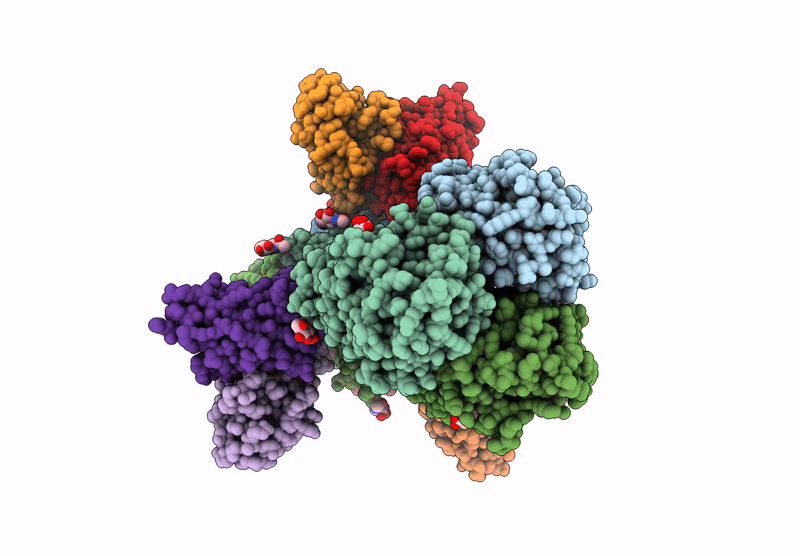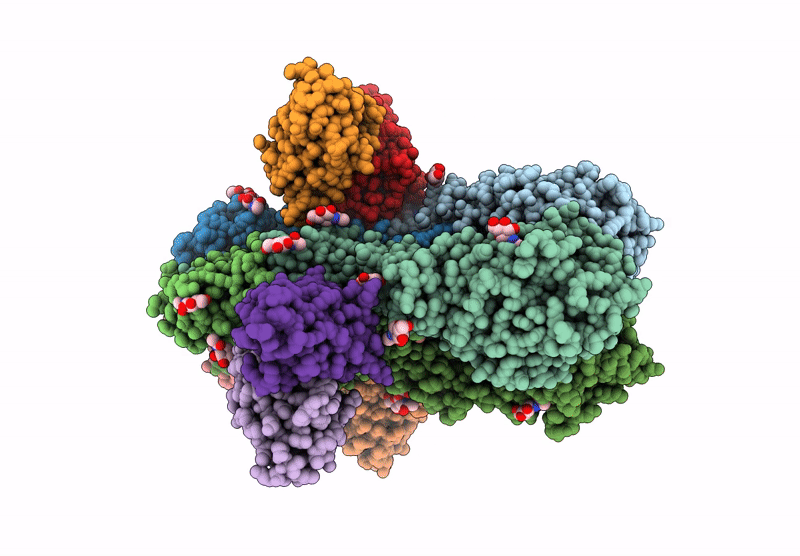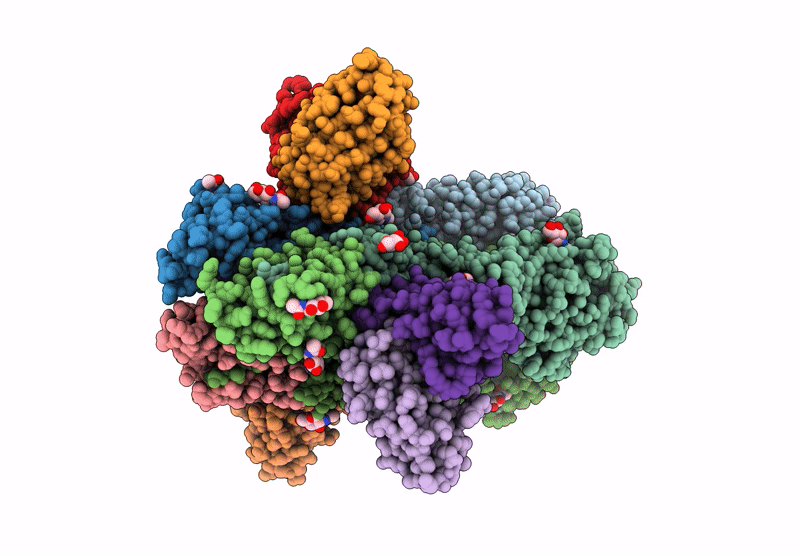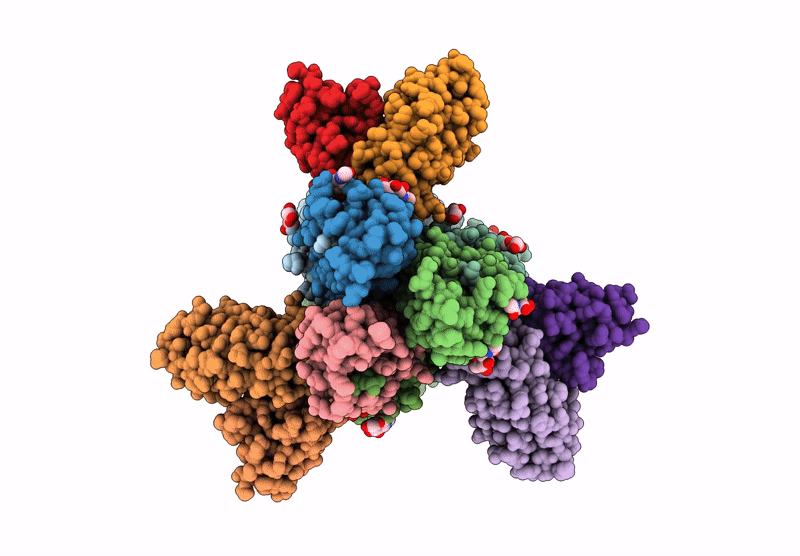
Hemagglutinin CA09 homotrimer bound to AEL31302/AEL31311 Fab
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Influenza A virus (A/California/01/2009(H1N1)) (Taxon ID: 666207)
Influenza A virus (A/California/01/2009(H1N1)) (Taxon ID: 666207)
Host Organism:
Method Details:
Experimental Method:
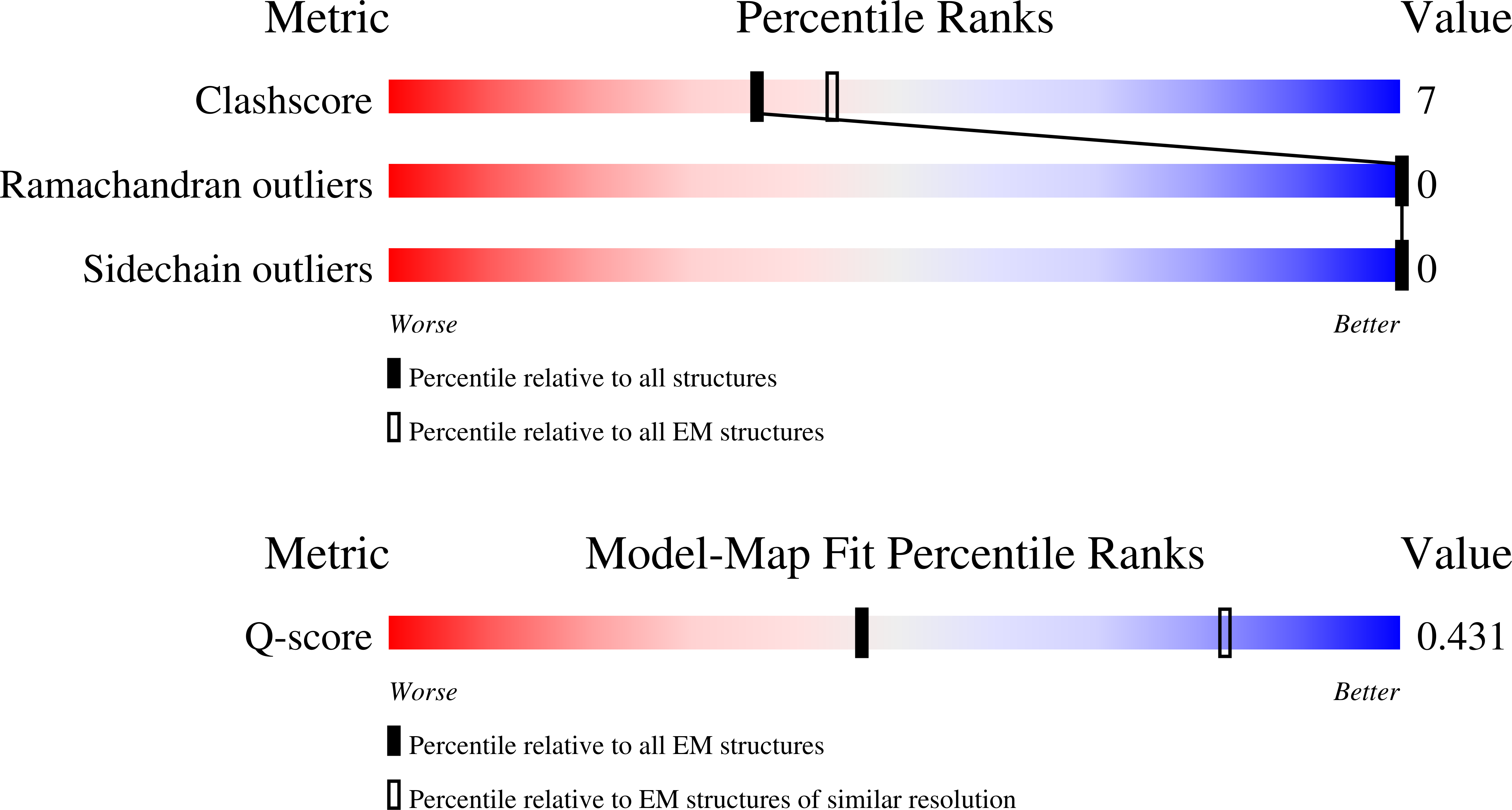
Resolution:
4.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE