Deposition Date
2025-01-16
Release Date
2025-05-21
Last Version Date
2025-05-28
Entry Detail
PDB ID:
9MVY
Keywords:
Title:
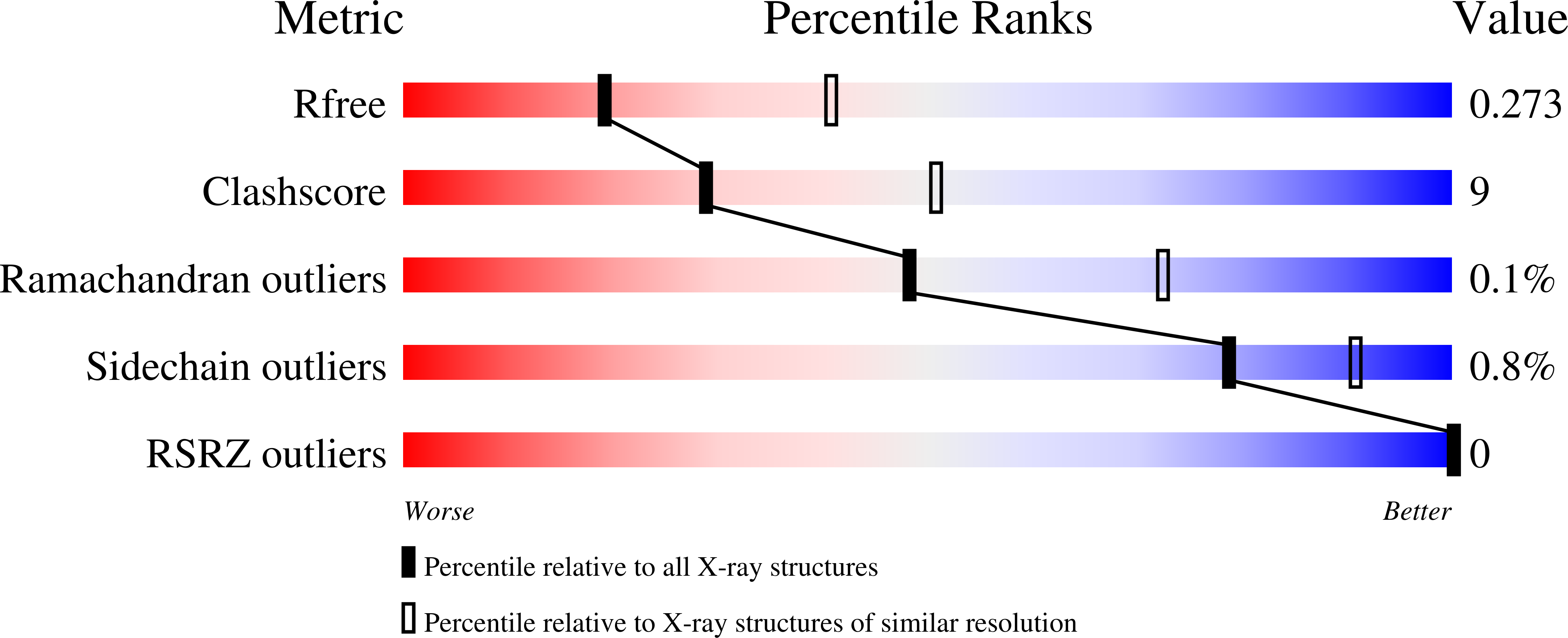
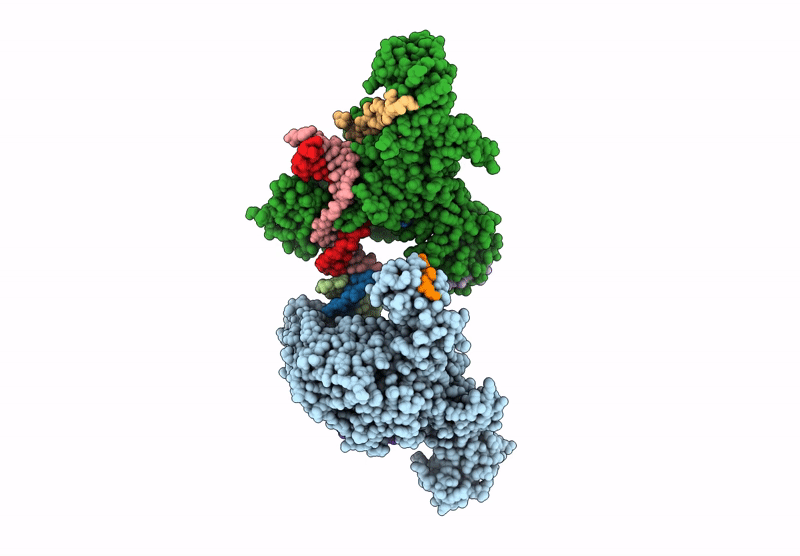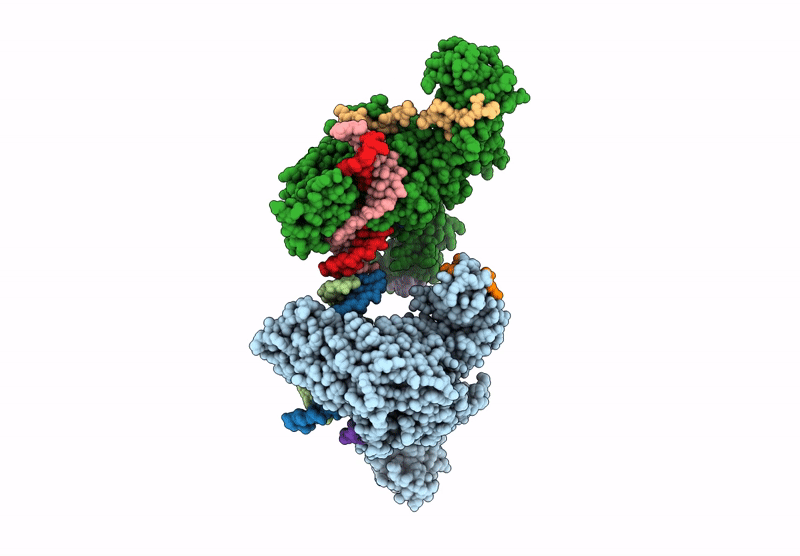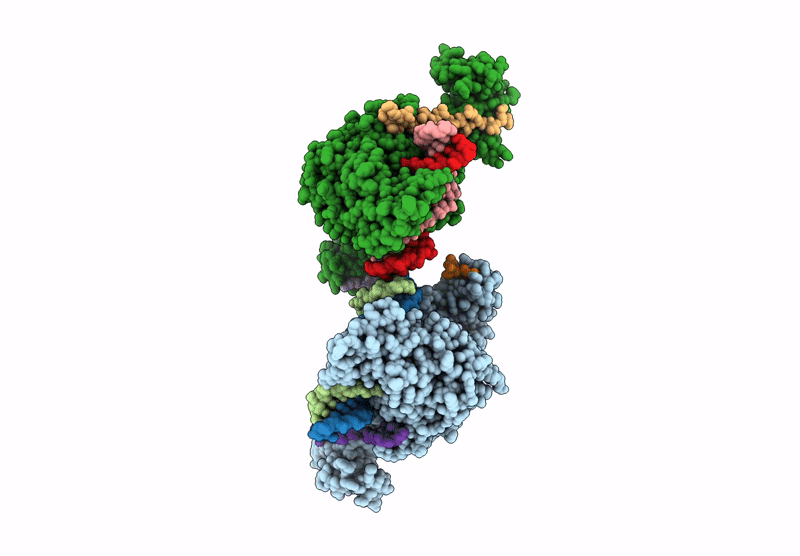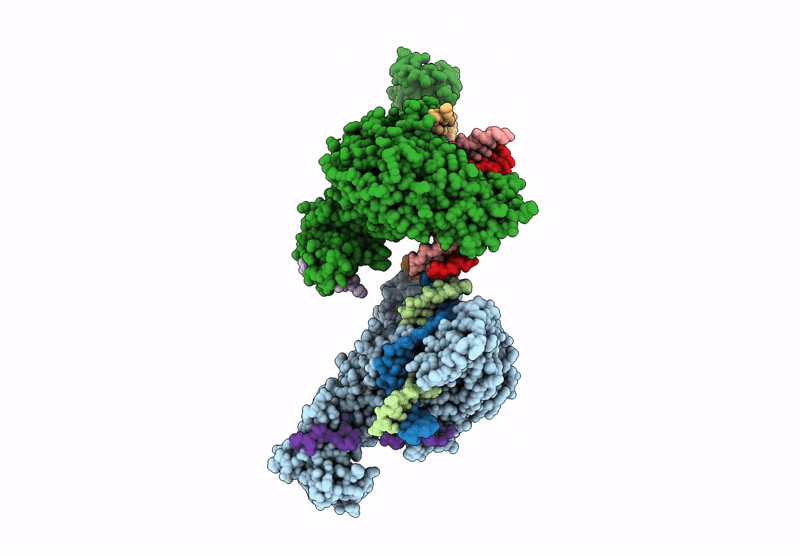
Crystal structure of ZMET2 in complex with unmethylated CTG DNA and a histone H3Kc9me2 peptide
Biological Source:
Source Organism:
Zea mays (Taxon ID: 4577)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.71 Å
R-Value Free:
0.27
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 1 21 1