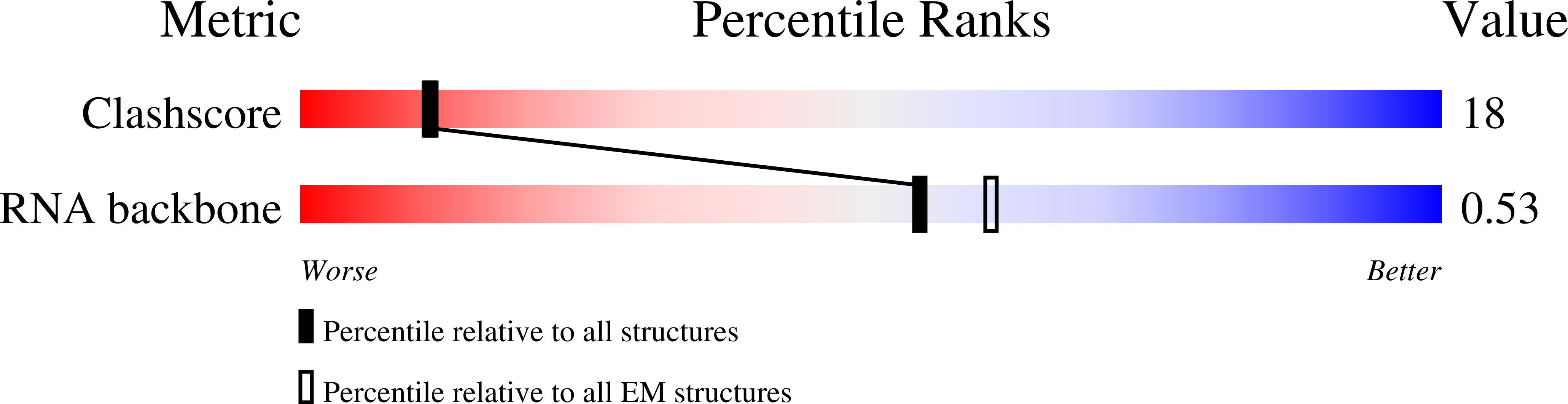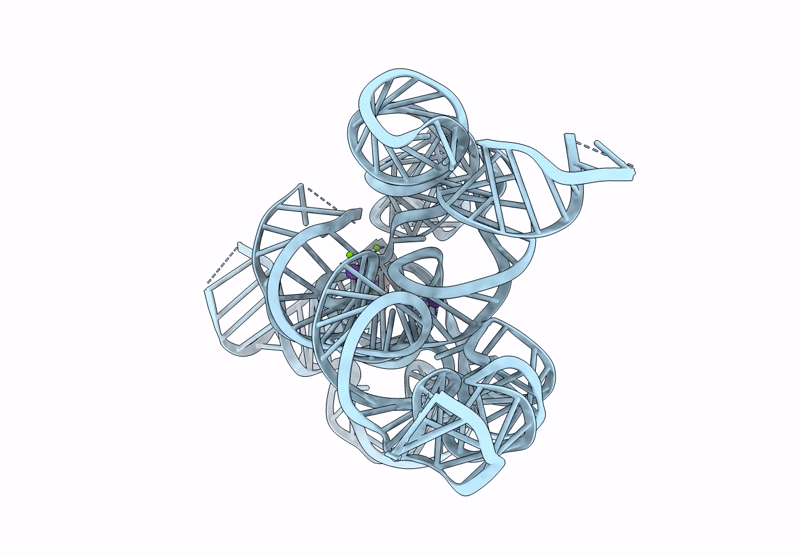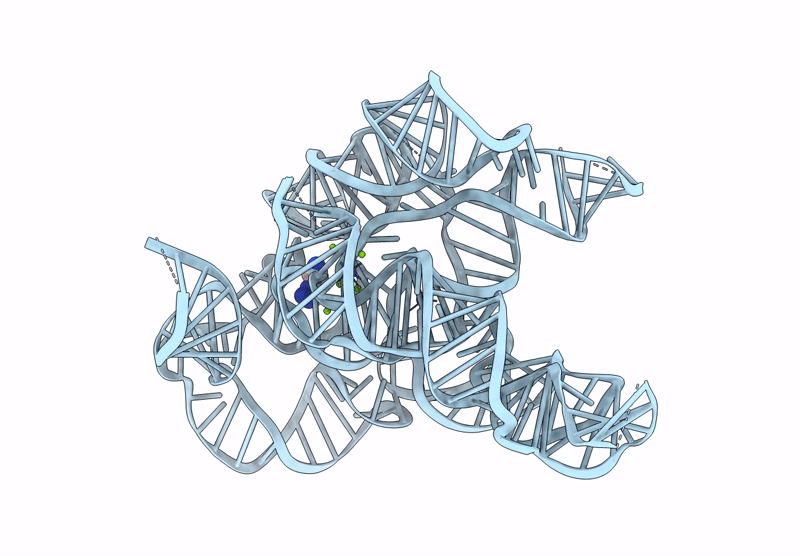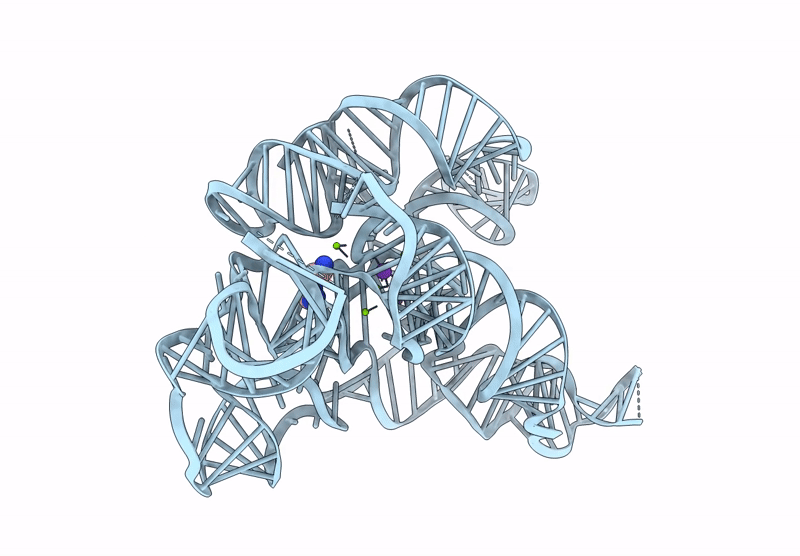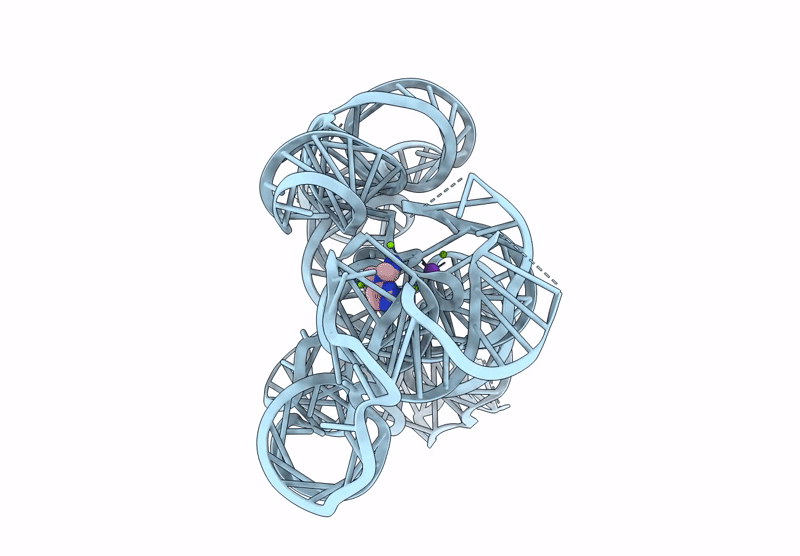
Deposition Date
2025-01-06
Release Date
2025-04-30
Last Version Date
2025-05-28
Entry Detail
PDB ID:
9MQT
Keywords:
Title:
CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Magnesium Condition
Biological Source:
Source Organism(s):
Candida albicans (Taxon ID: 5476)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.43 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE