Deposition Date
2025-03-07
Release Date
2025-11-12
Last Version Date
2025-11-12
Entry Detail
PDB ID:
9M6O
Keywords:
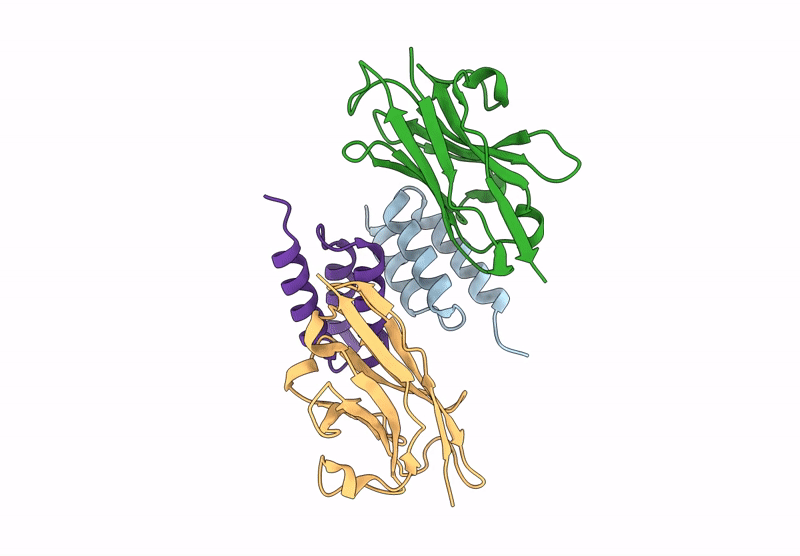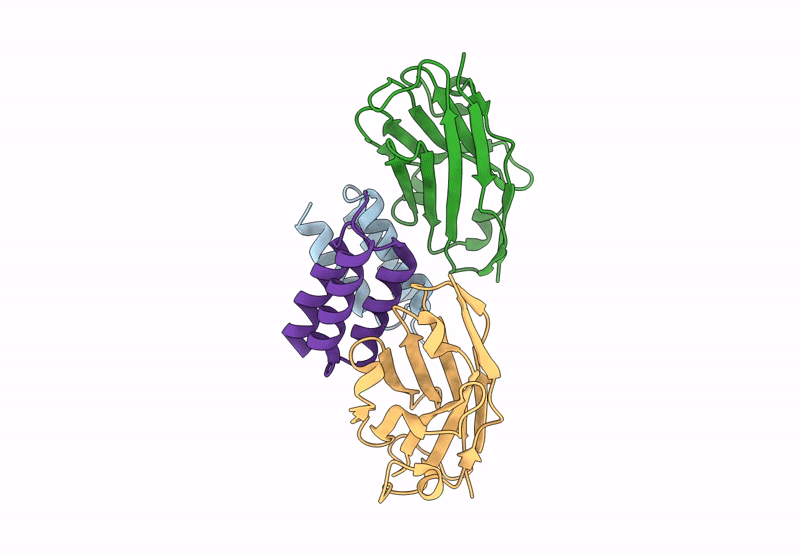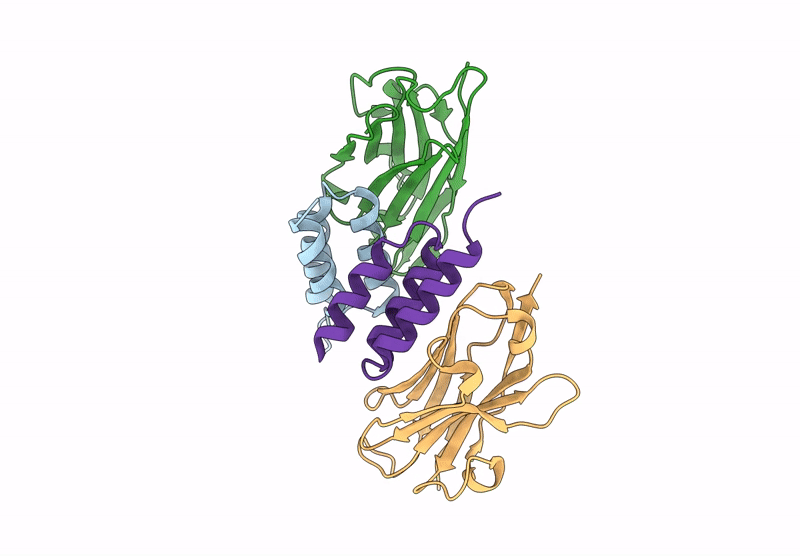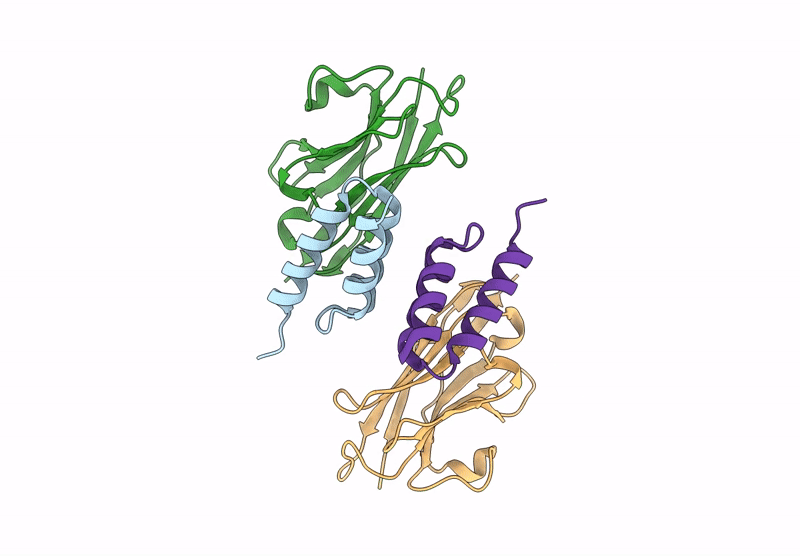
Title:
Crystal structure of S. aureus protein A bound to a camelid single-domain antibody
Biological Source:
Source Organism:
Staphylococcus aureus (strain NCTC 8325 / PS 47) (Taxon ID: 93061)
Camelus dromedarius (Taxon ID: 9838)
Camelus dromedarius (Taxon ID: 9838)
Host Organism:
Method Details:
Experimental Method:
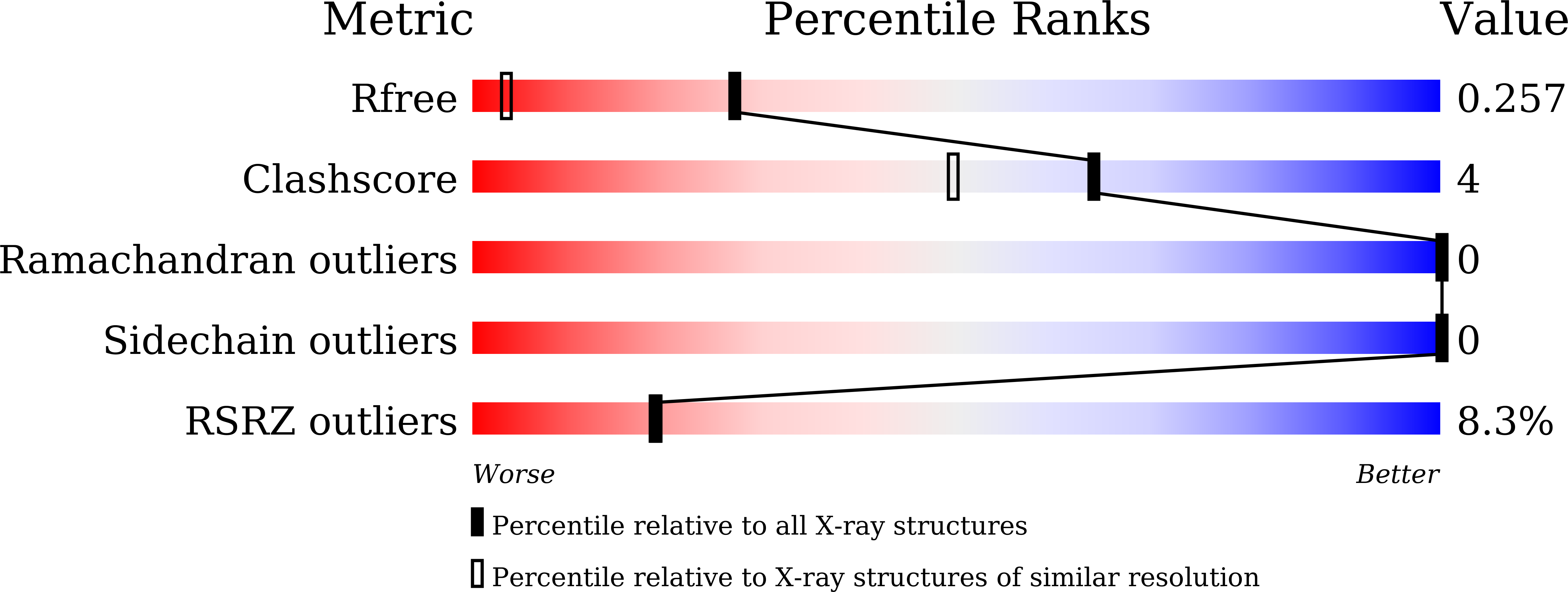
Resolution:
1.49 Å
R-Value Free:
0.25
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 21 21 21