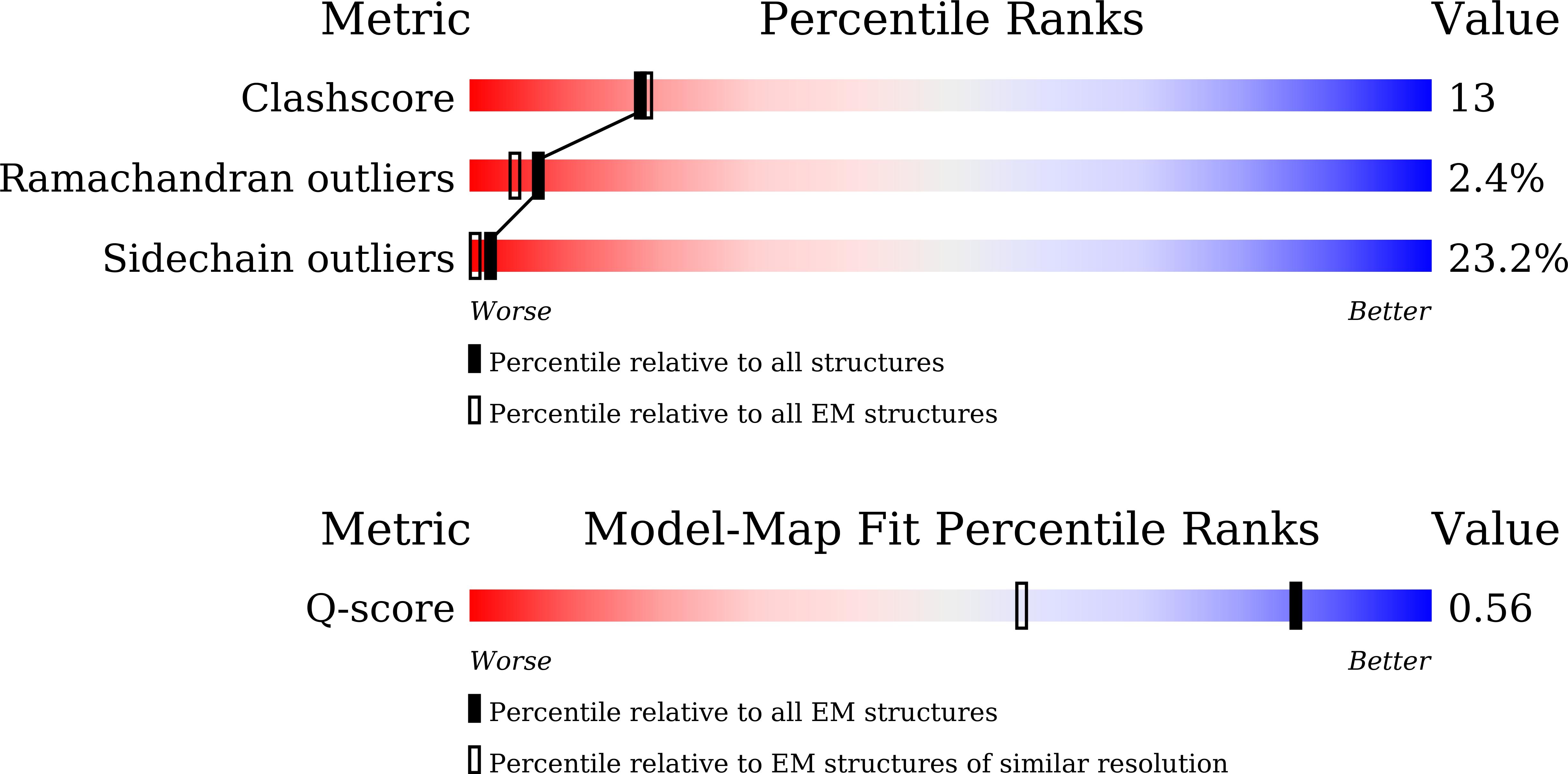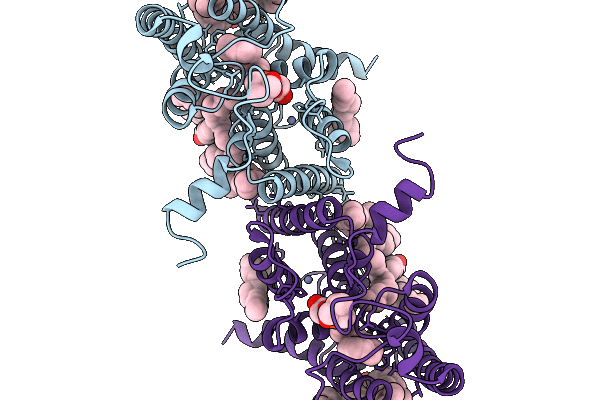
Deposition Date
2025-02-13
Release Date
2026-01-07
Last Version Date
2026-02-18
Entry Detail
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE