Deposition Date
2024-11-24
Release Date
2024-12-11
Last Version Date
2025-08-20
Entry Detail
PDB ID:
9KPY
Keywords:
Title:
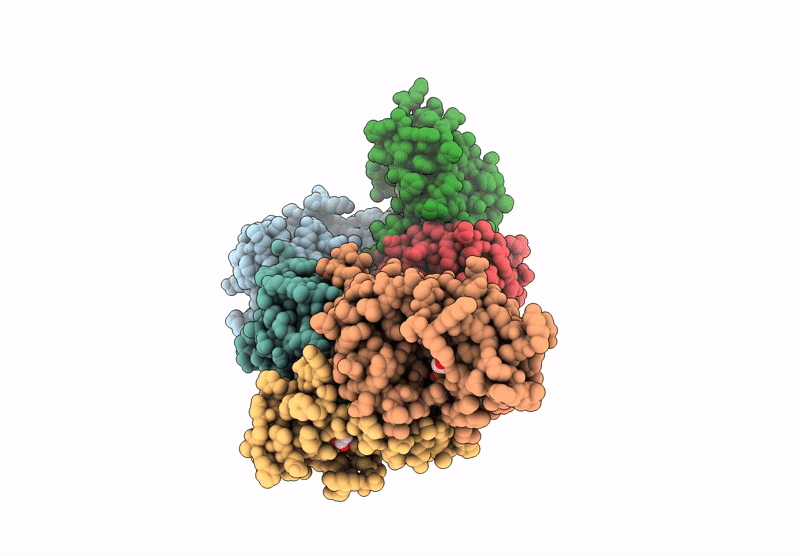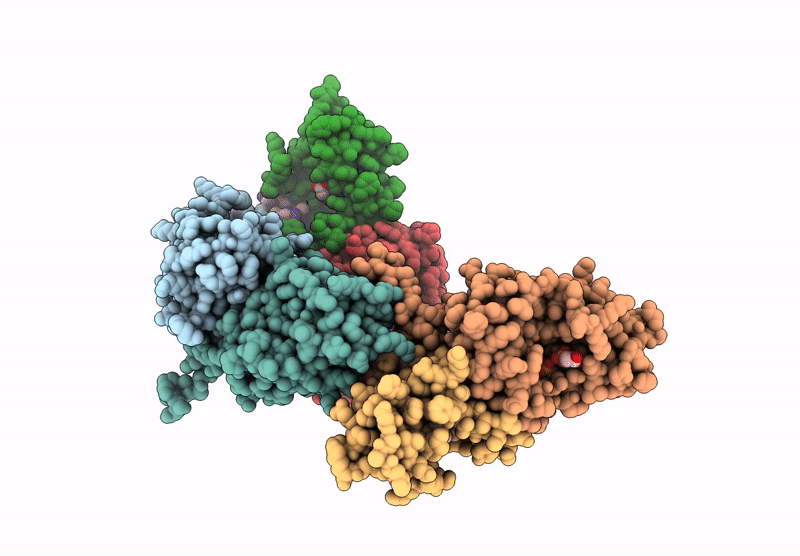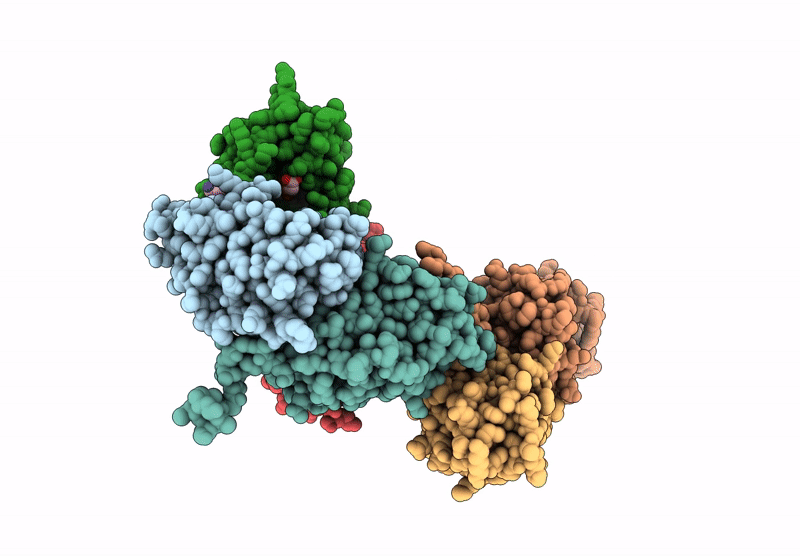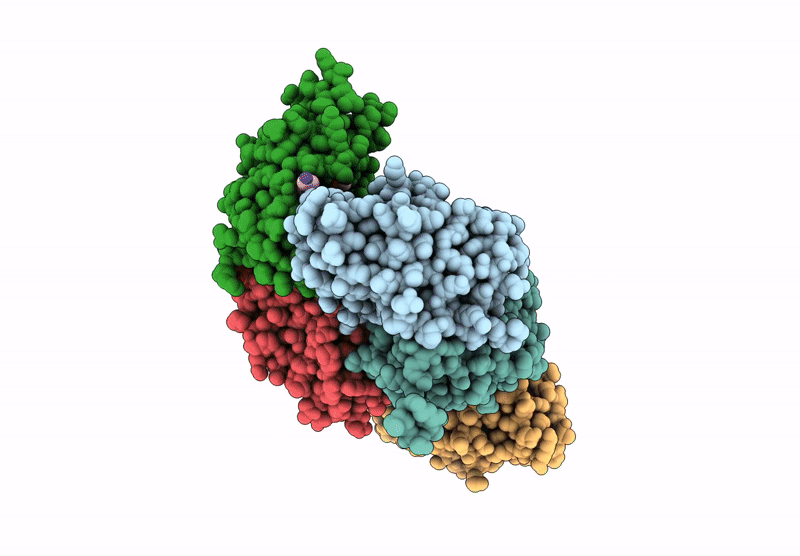
Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.20 A resolution.
Biological Source:
Source Organism(s):
Enterobacter sp. 638 (Taxon ID: 399742)
Expression System(s):
Method Details:
Experimental Method:
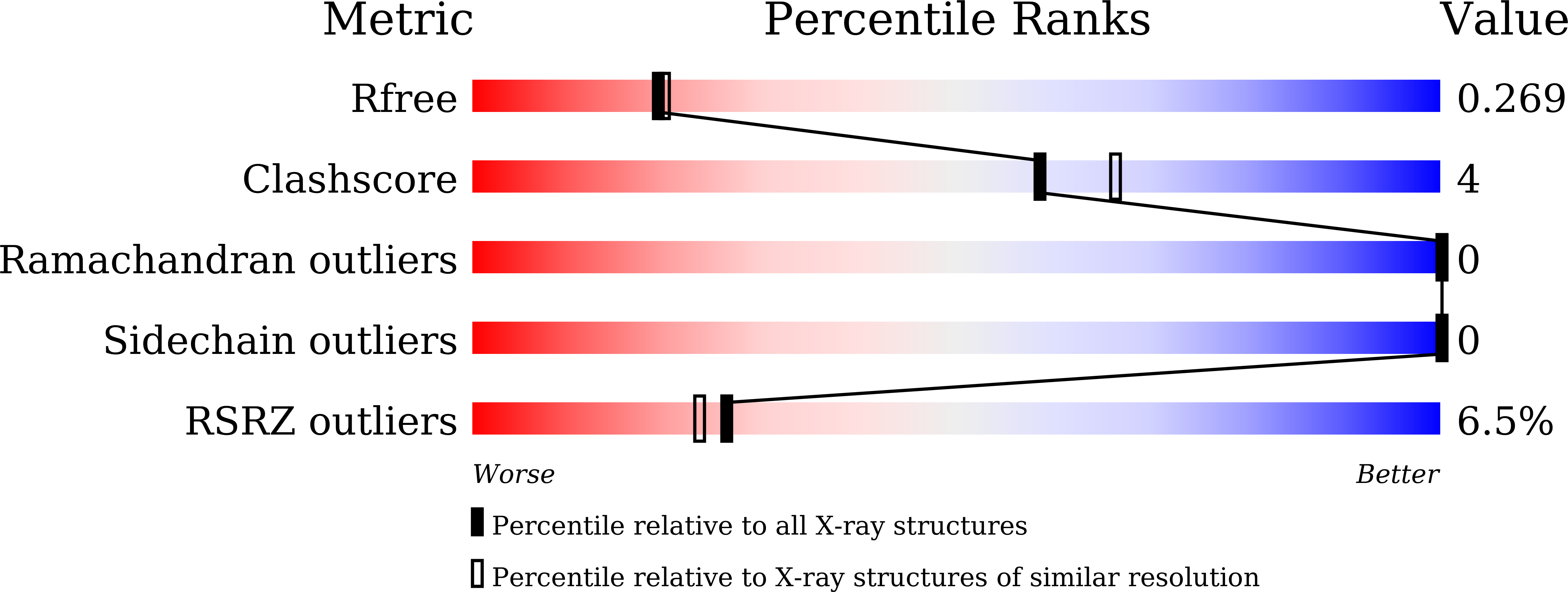
Resolution:
2.20 Å
R-Value Free:
0.26
R-Value Work:
0.20
Space Group:
C 1 2 1