Deposition Date
2024-11-24
Release Date
2025-11-26
Last Version Date
2026-01-14
Entry Detail
PDB ID:
9KPR
Keywords:
Title:
Crystal structure of a allulose transcriptional regulator from Agrobacterium fabrum
Biological Source:
Source Organism(s):
Agrobacterium fabrum str. C58 (Taxon ID: 176299)
Expression System(s):
Method Details:
Experimental Method:
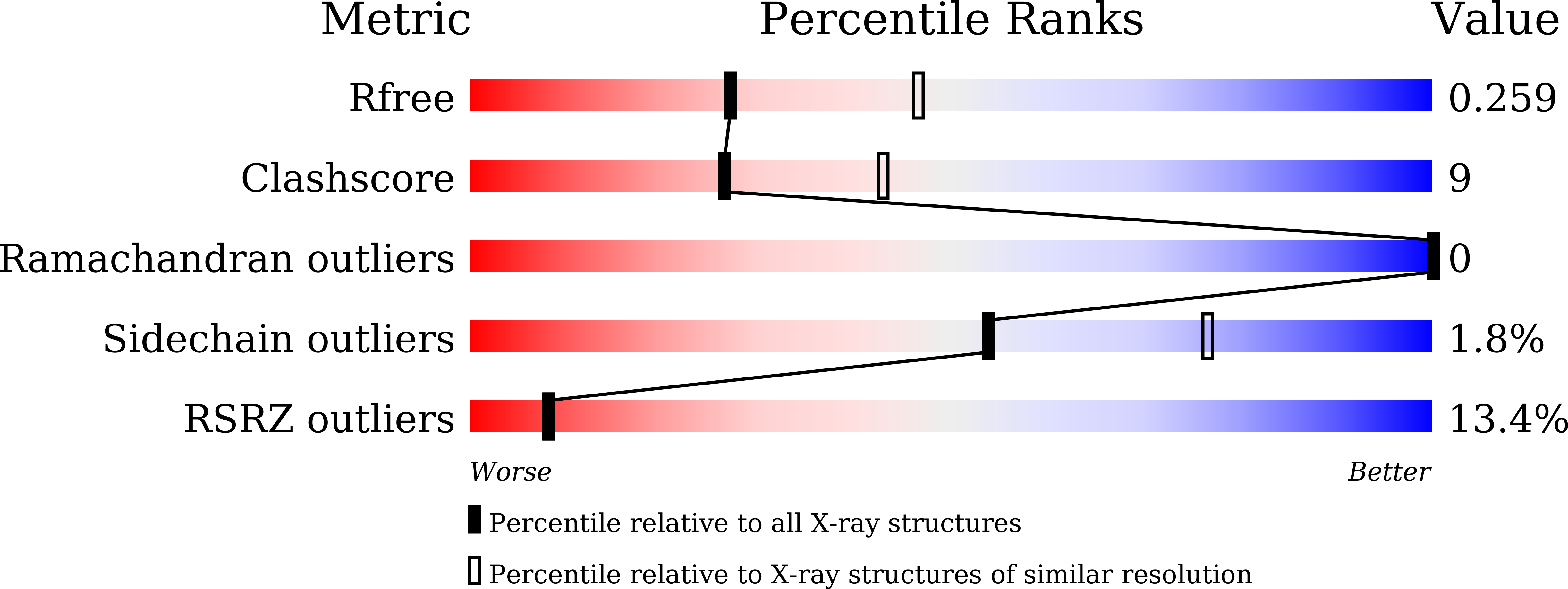
Resolution:
2.53 Å
R-Value Free:
0.26
R-Value Work:
0.23
Space Group:
P 41 21 2