Deposition Date
2024-09-27
Release Date
2025-08-06
Last Version Date
2025-08-06
Entry Detail
PDB ID:
9JQA
Keywords:
Title:
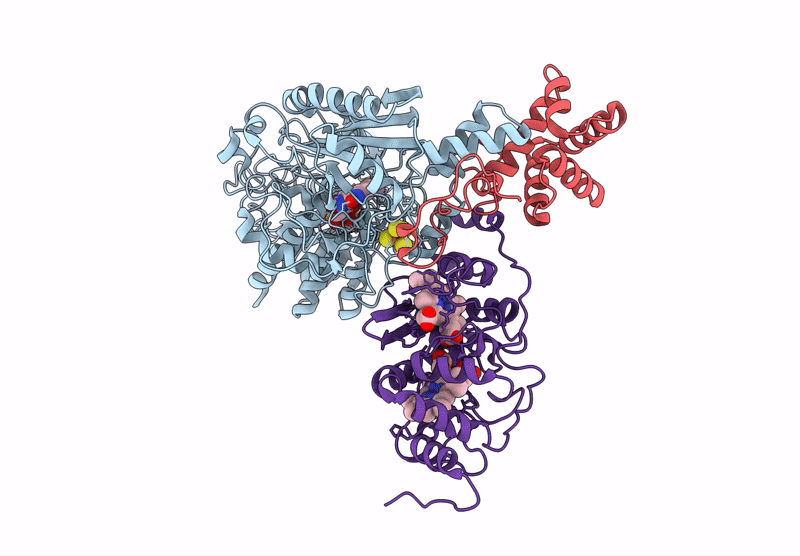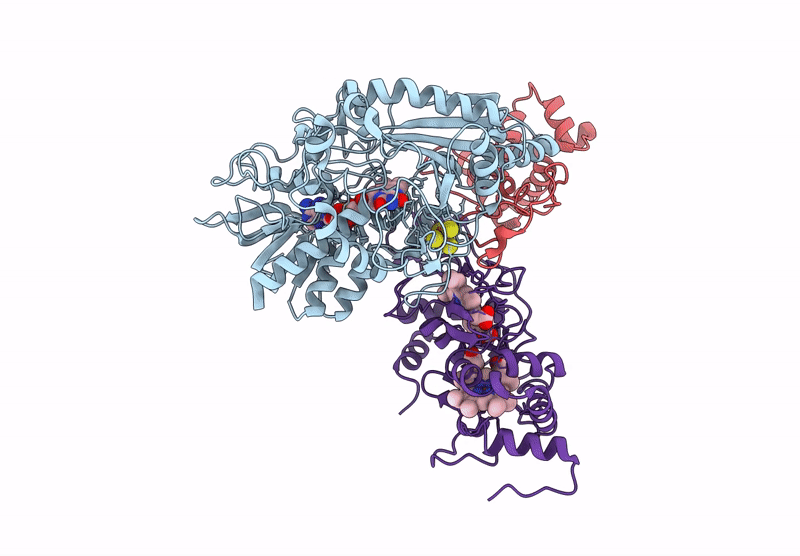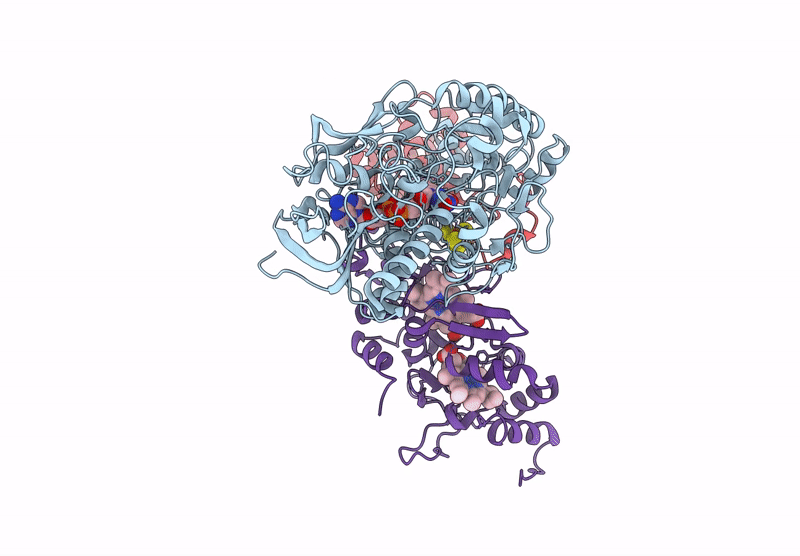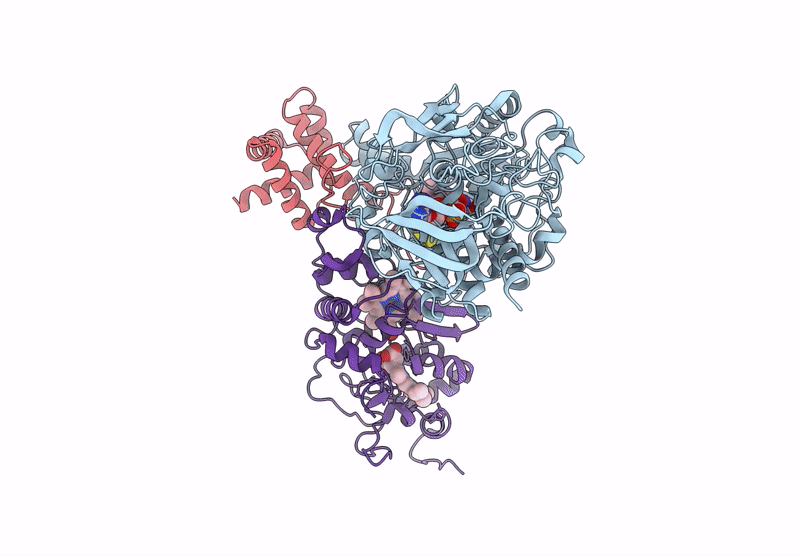
Cryo-EM Structure of Fructose Dehydrogenase Variant from Gluconobacter japonicus Truncating Heme 1c and C-Terminal Hydrophobic Regions
Biological Source:
Source Organism:
Gluconobacter japonicus (Taxon ID: 376620)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.15 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


