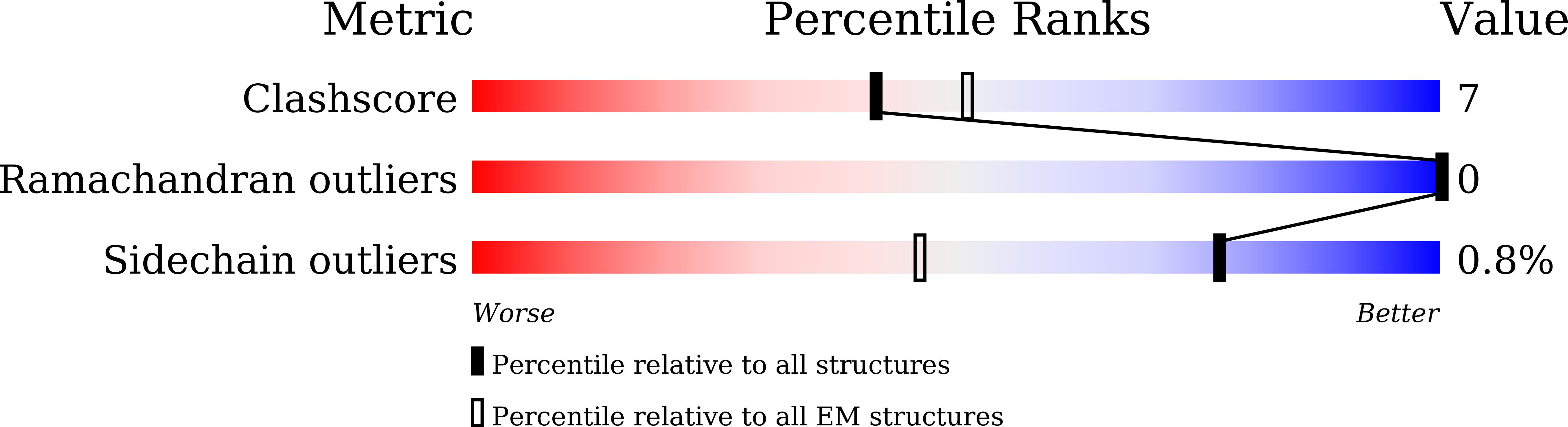
Deposition Date
2024-09-13
Release Date
2025-08-06
Last Version Date
2025-08-13
Entry Detail
PDB ID:
9JJ9
Keywords:
Title:
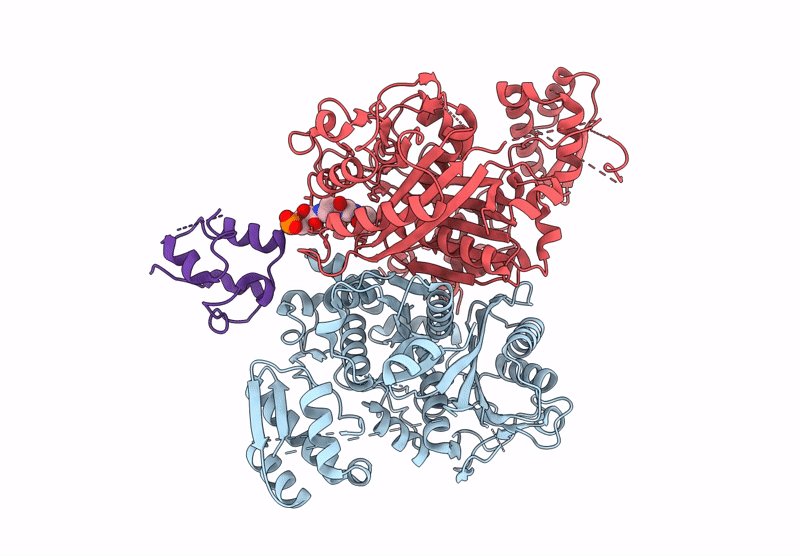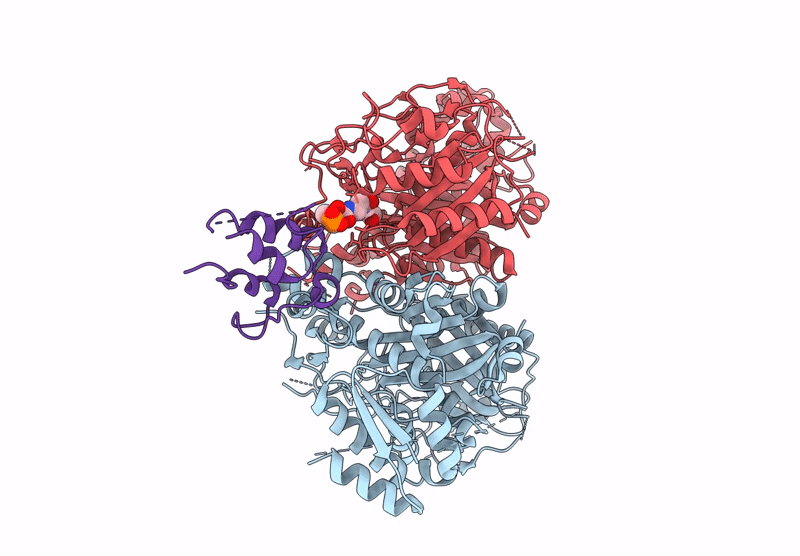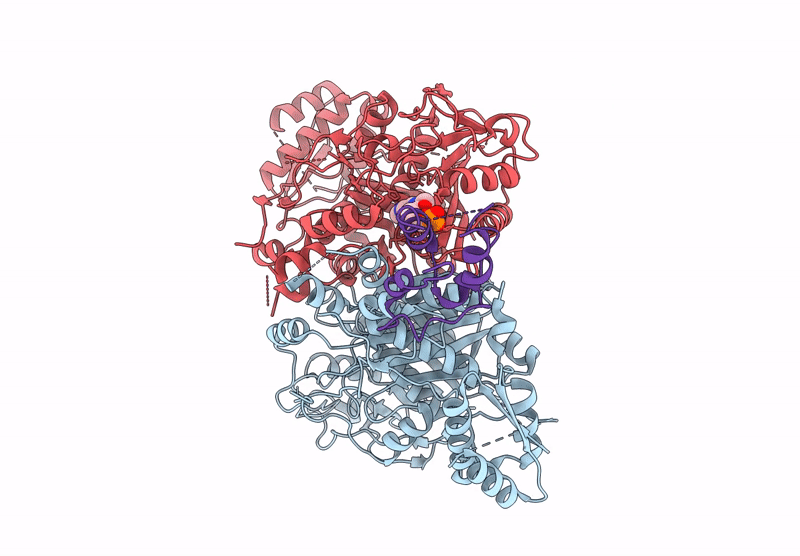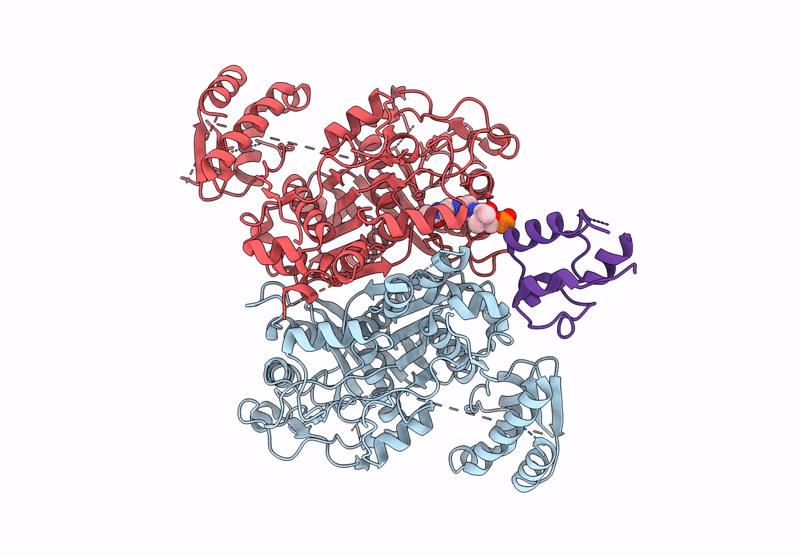
Class 3 state of the GfsA KSQ-ancestralAT chimeric didomain in complex with the GfsA ACP domain
Biological Source:
Source Organism:
Streptomyces graminofaciens (Taxon ID: 68212)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.71 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE