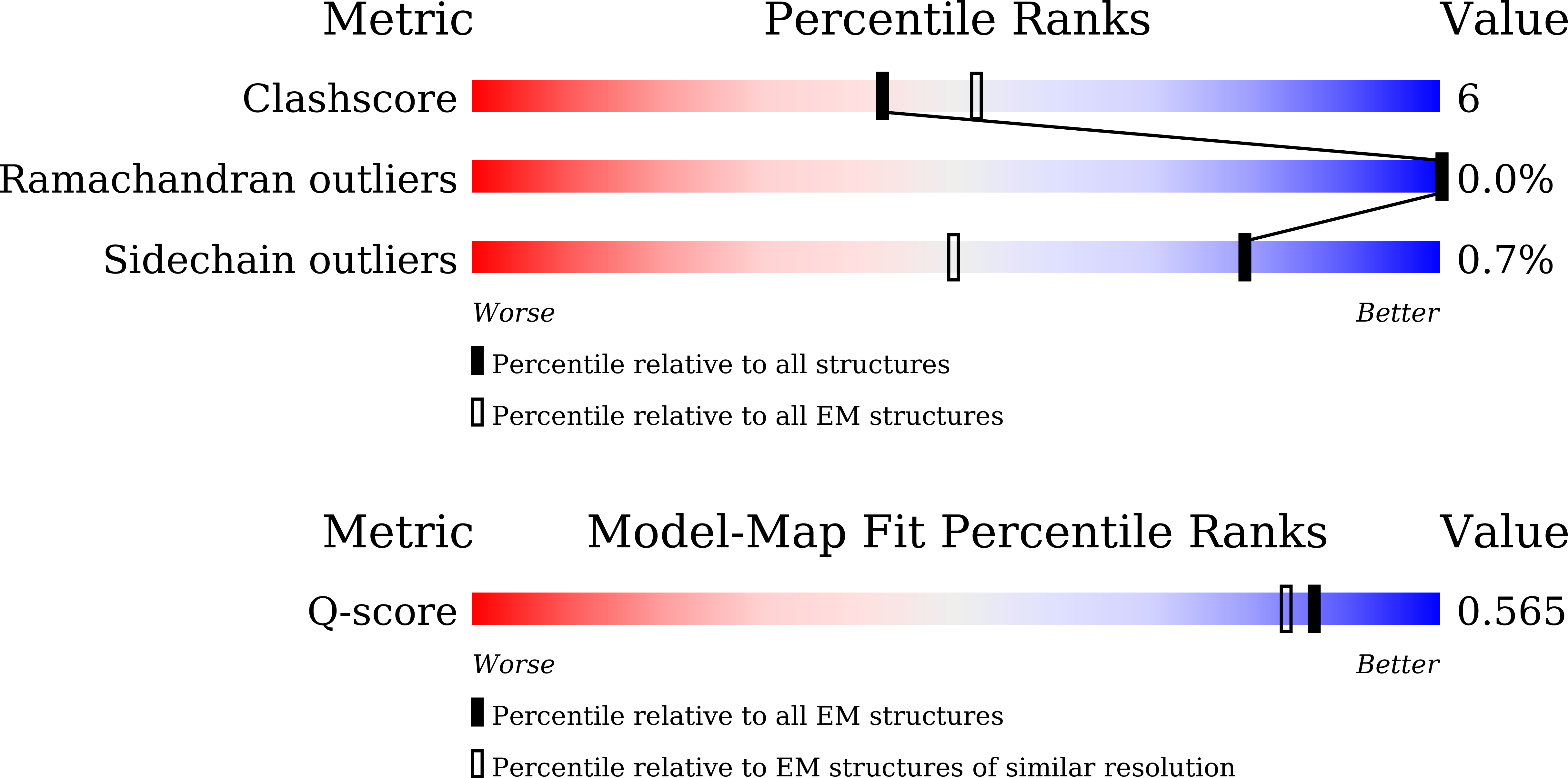
Deposition Date
2024-08-17
Release Date
2025-11-19
Last Version Date
2026-02-04
Entry Detail
PDB ID:
9J6W
Keywords:
Title:
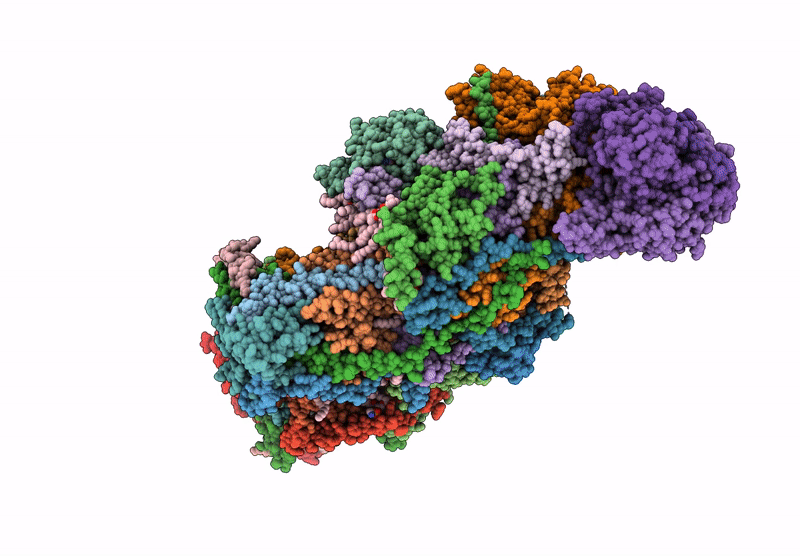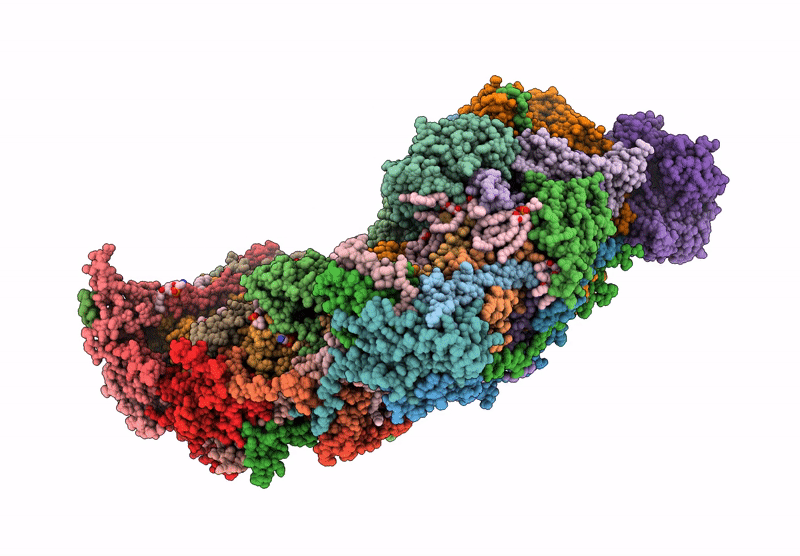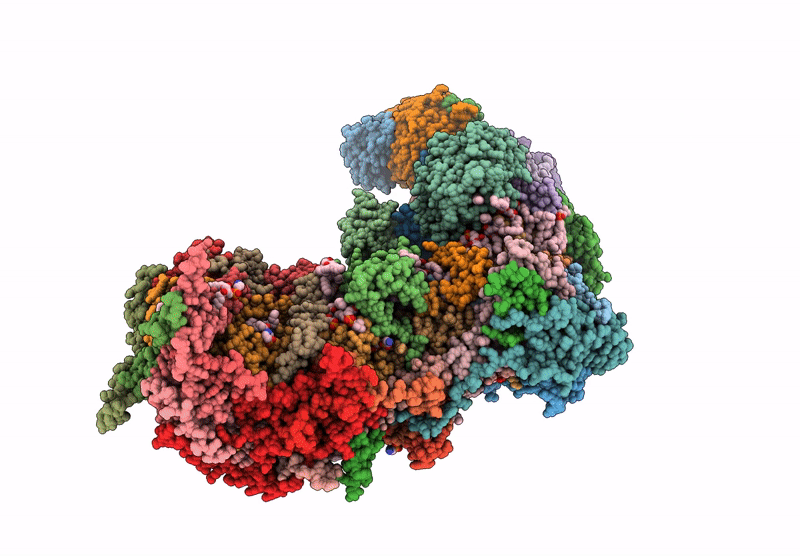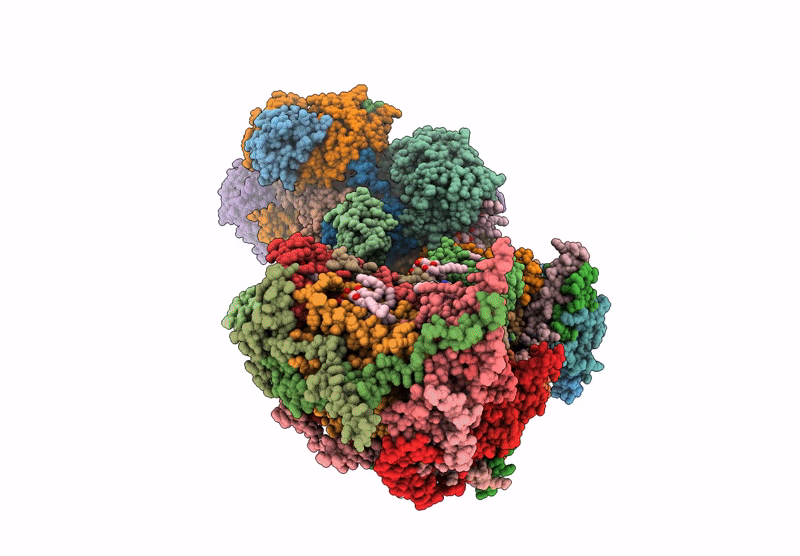
Complex I from respirasome closed state 1 bound by metformin and CoQ10 (SC-MetC1-v)
Biological Source:
Source Organism(s):
Sus scrofa (Taxon ID: 9823)
Method Details:
Experimental Method:
Resolution:
2.99 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE