Deposition Date
2025-01-24
Release Date
2025-10-22
Last Version Date
2025-11-19
Entry Detail
PDB ID:
9I3U
Keywords:
Title:
Cryo-EM structure of the AGR2 dimer in complex with the monomeric IRE1beta luminal domain
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
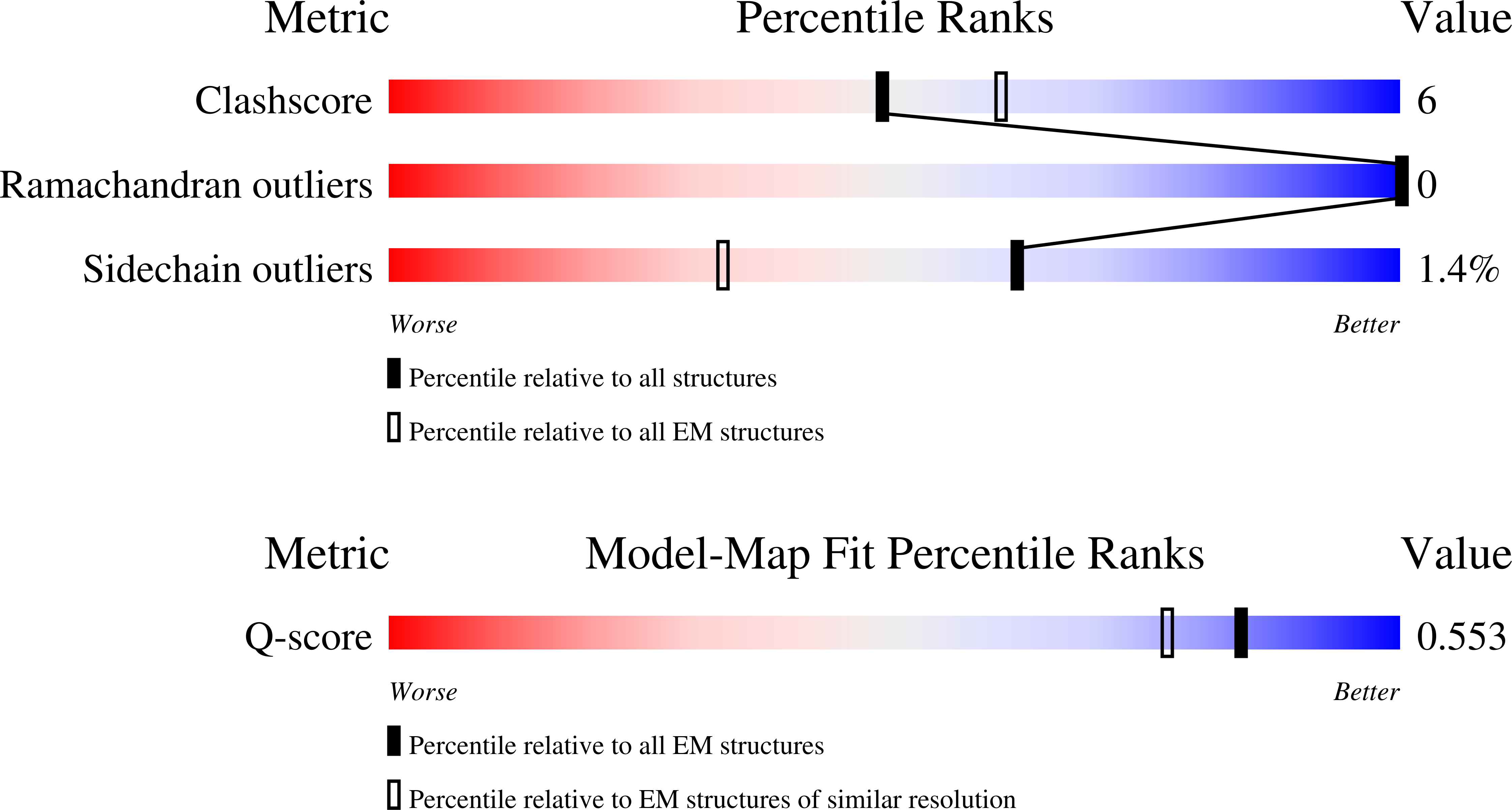
Resolution:
2.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE