Deposition Date
2025-01-10
Release Date
2025-07-16
Last Version Date
2025-07-30
Entry Detail
PDB ID:
9HYQ
Keywords:
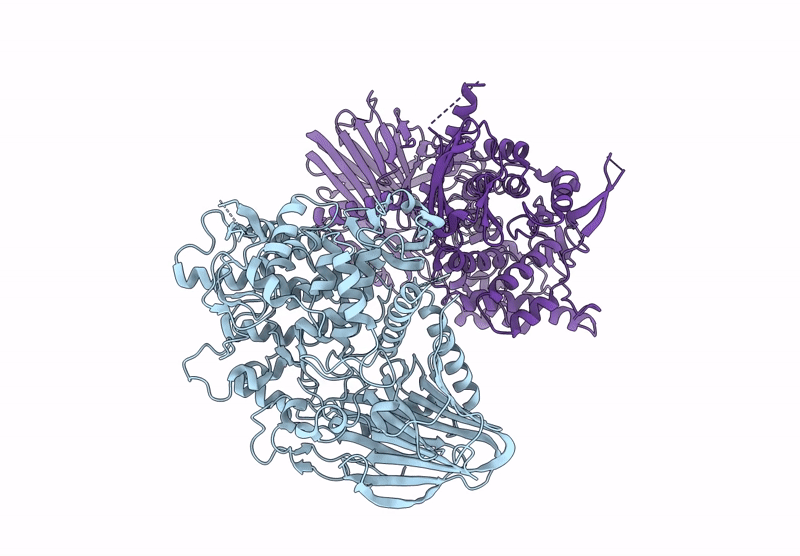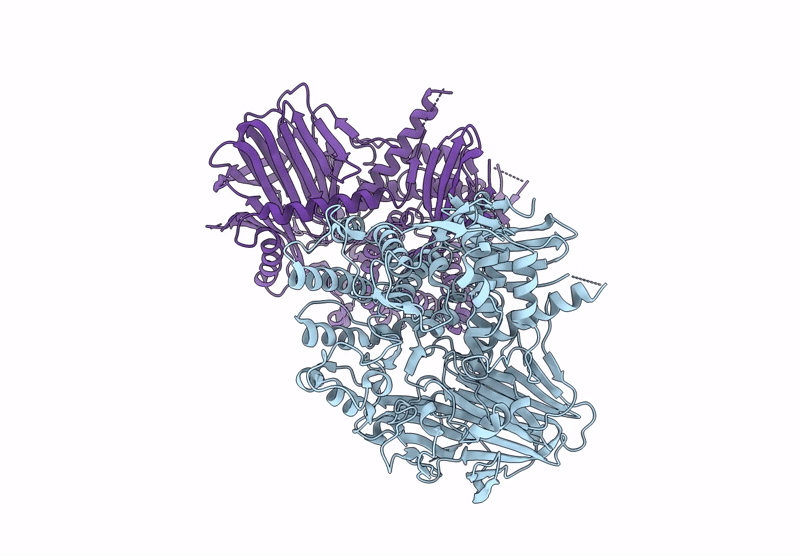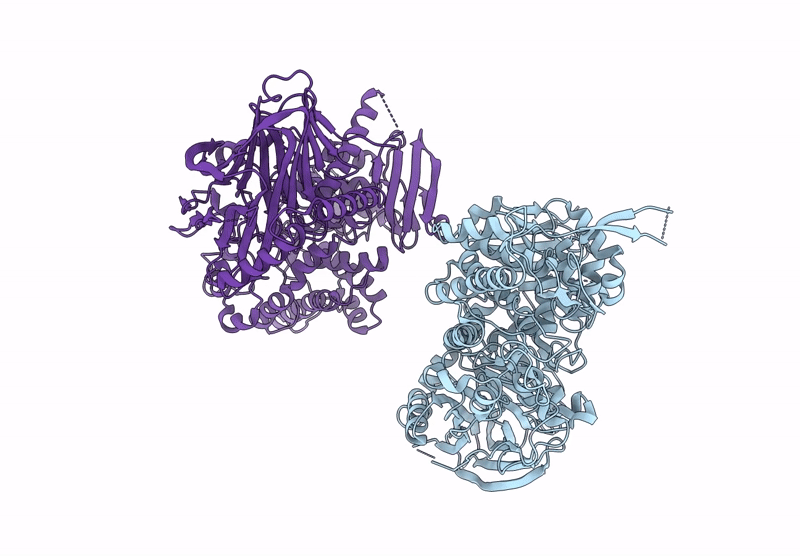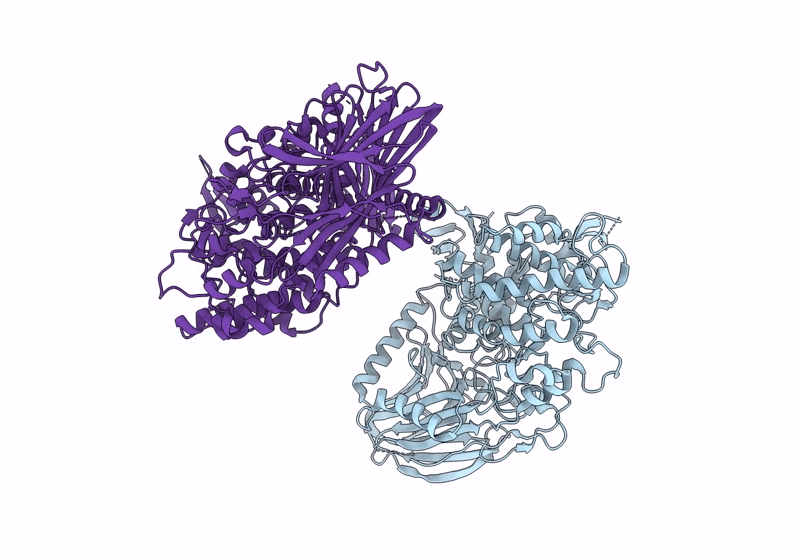
Title:
BT984 a GH139 rhamnogalacturonan II exo-a-1,2-(2-Omethyl)-fucosidase
Biological Source:
Source Organism(s):
Bacteroides thetaiotaomicron VPI-5482 (Taxon ID: 226186)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.70 Å
R-Value Free:
0.25
R-Value Work:
0.21
Space Group:
P 21 21 21


