Deposition Date
2024-12-11
Release Date
2025-05-07
Last Version Date
2025-05-21
Entry Detail
PDB ID:
9HNO
Keywords:
Title:
Structure of the (6-4) photolyase of Caulobacter crescentus in its iron sulfur cluster oxidized state determined by serial femtosecond crystallography
Biological Source:
Source Organism:
Caulobacter vibrioides (Taxon ID: 155892)
Host Organism:
Method Details:
Experimental Method:
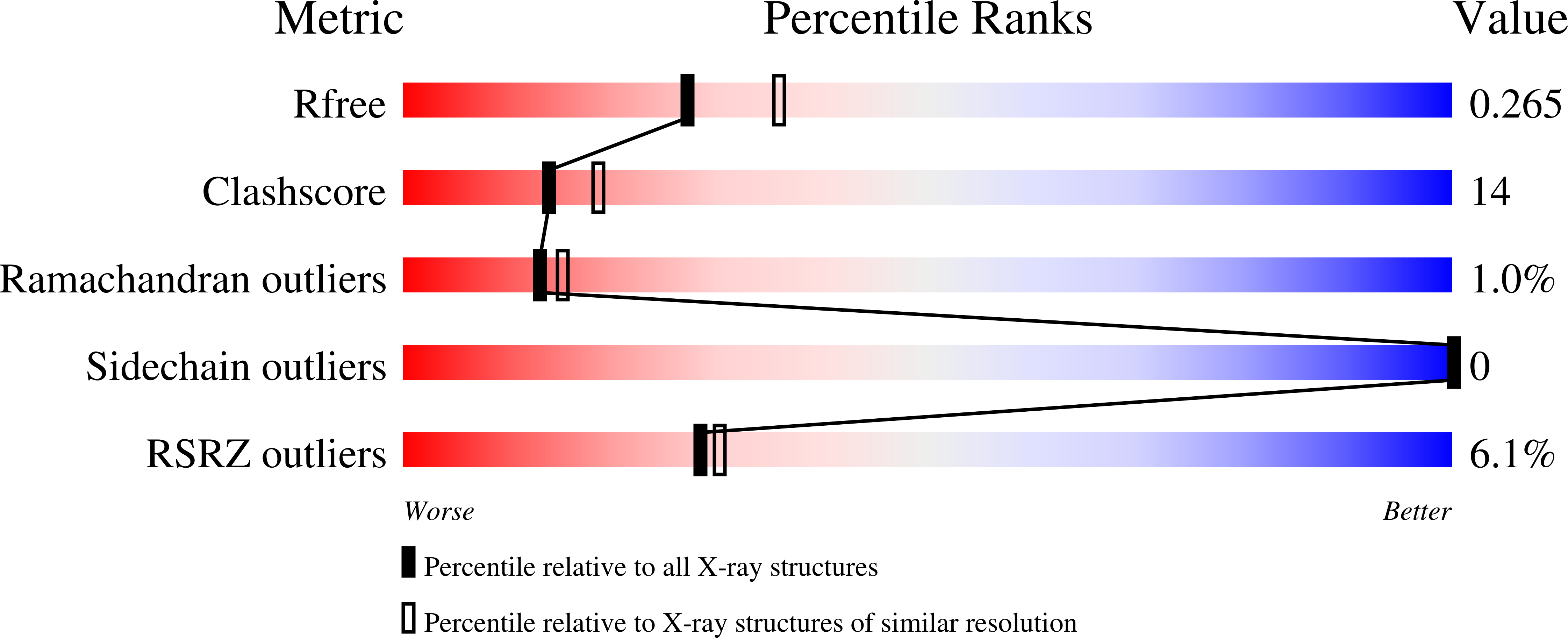
Resolution:
2.30 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 21 21