Deposition Date
2024-10-25
Release Date
2025-12-10
Last Version Date
2025-12-10
Entry Detail
PDB ID:
9H6Y
Keywords:
Title:
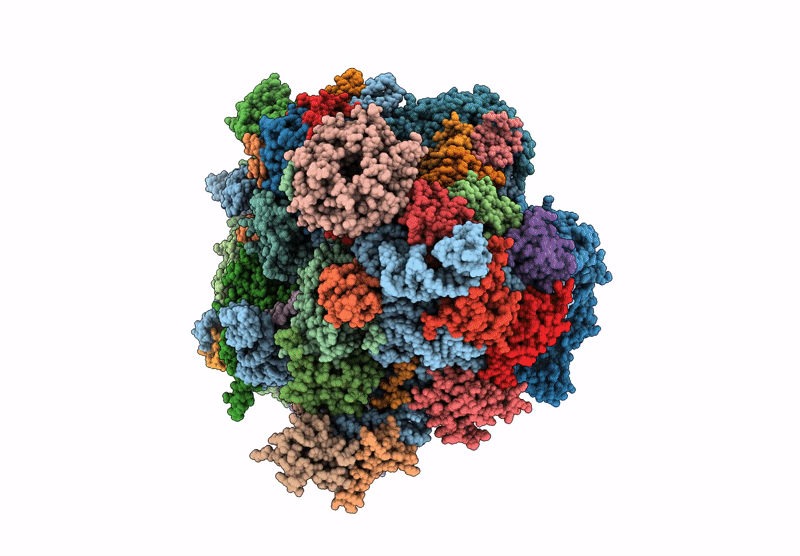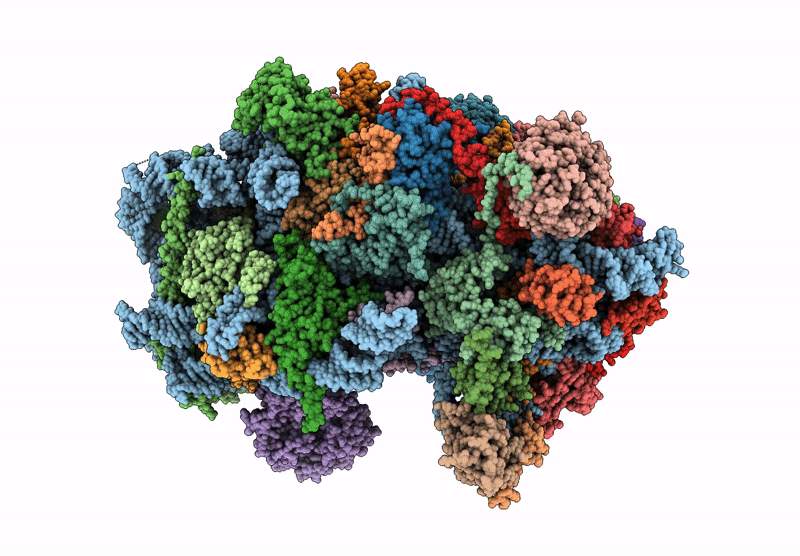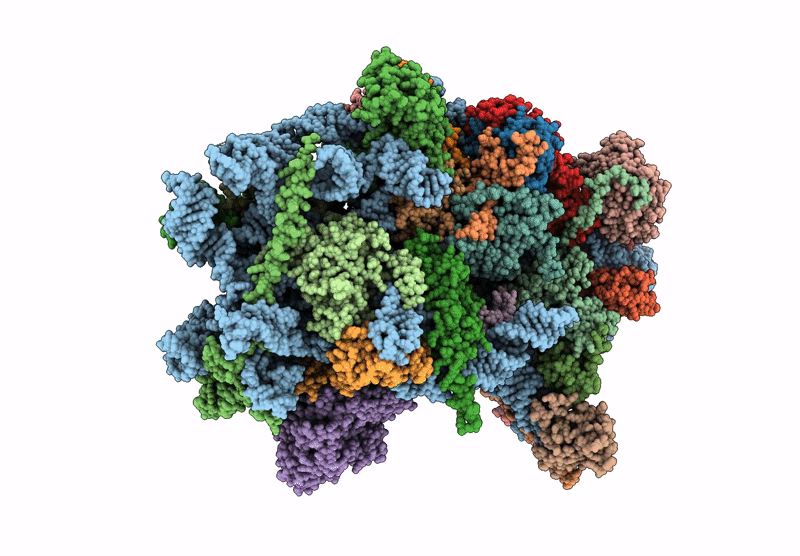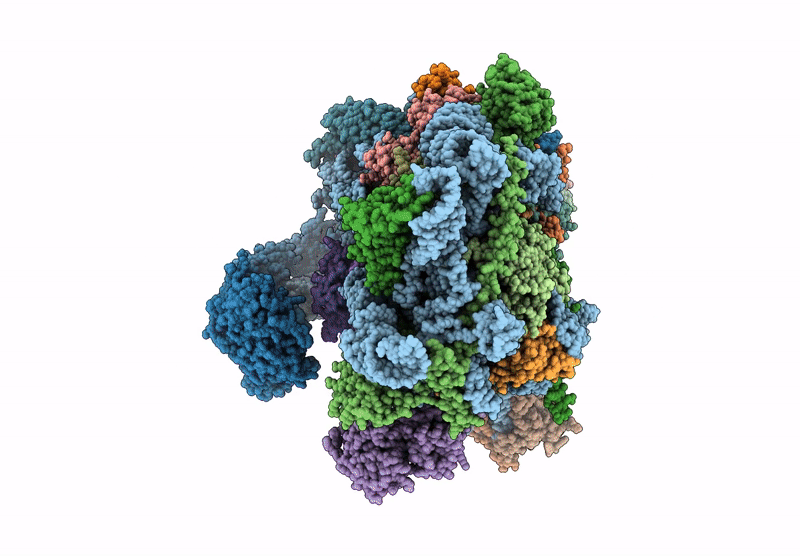
Late-stage 48S Initiation Complex (LS48S IC) guided by the trans-RNA
Biological Source:
Source Organism(s):
Oryctolagus cuniculus (Taxon ID: 9986)
Method Details:
Experimental Method:
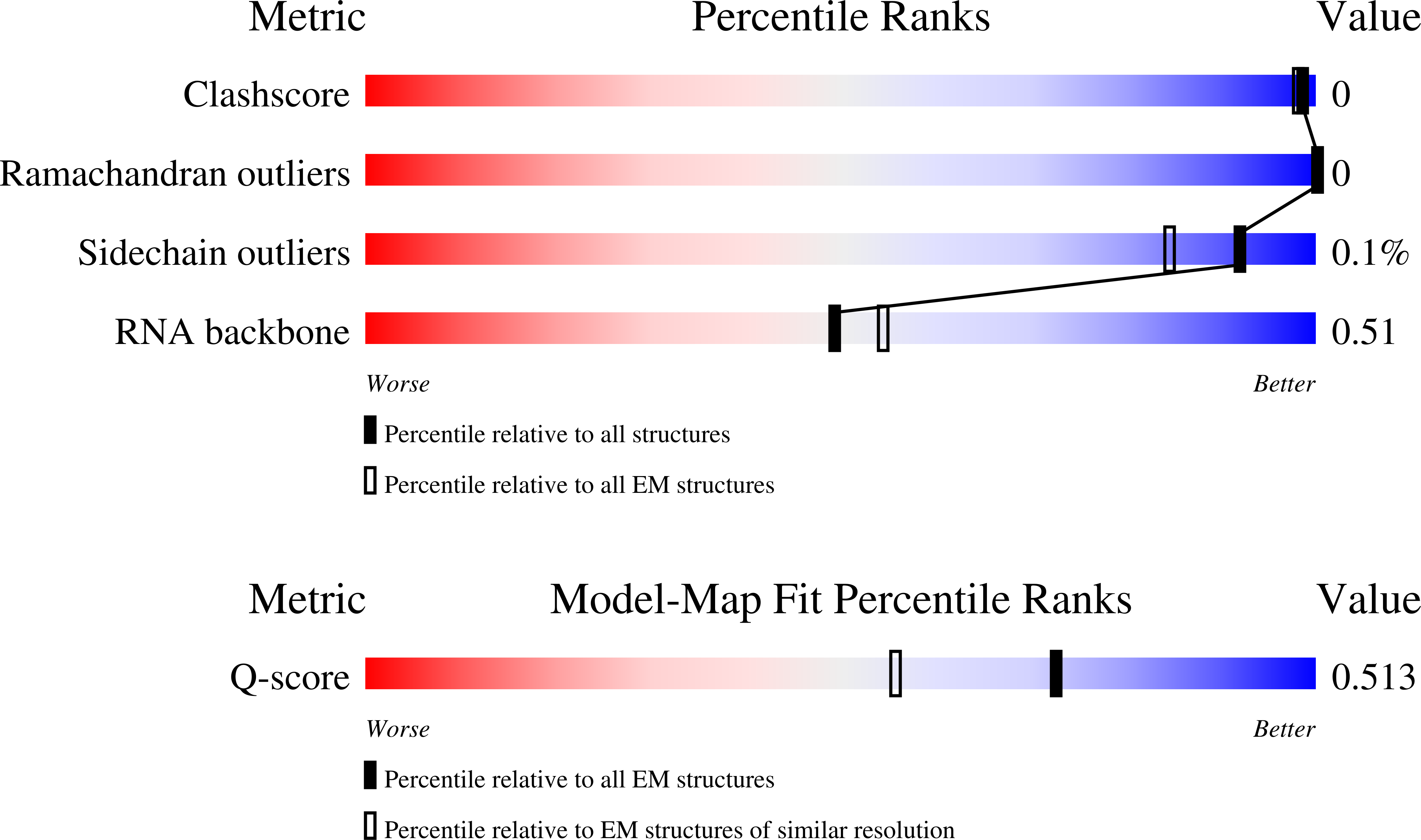
Resolution:
2.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE