Deposition Date
2024-10-15
Release Date
2025-06-18
Last Version Date
2025-07-16
Entry Detail
PDB ID:
9H2X
Keywords:
Title:
Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 7, a novel nanomolar A2A receptor antagonist from modern hit-finding with structure-guided de novo design
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Escherichia coli (Taxon ID: 562)
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
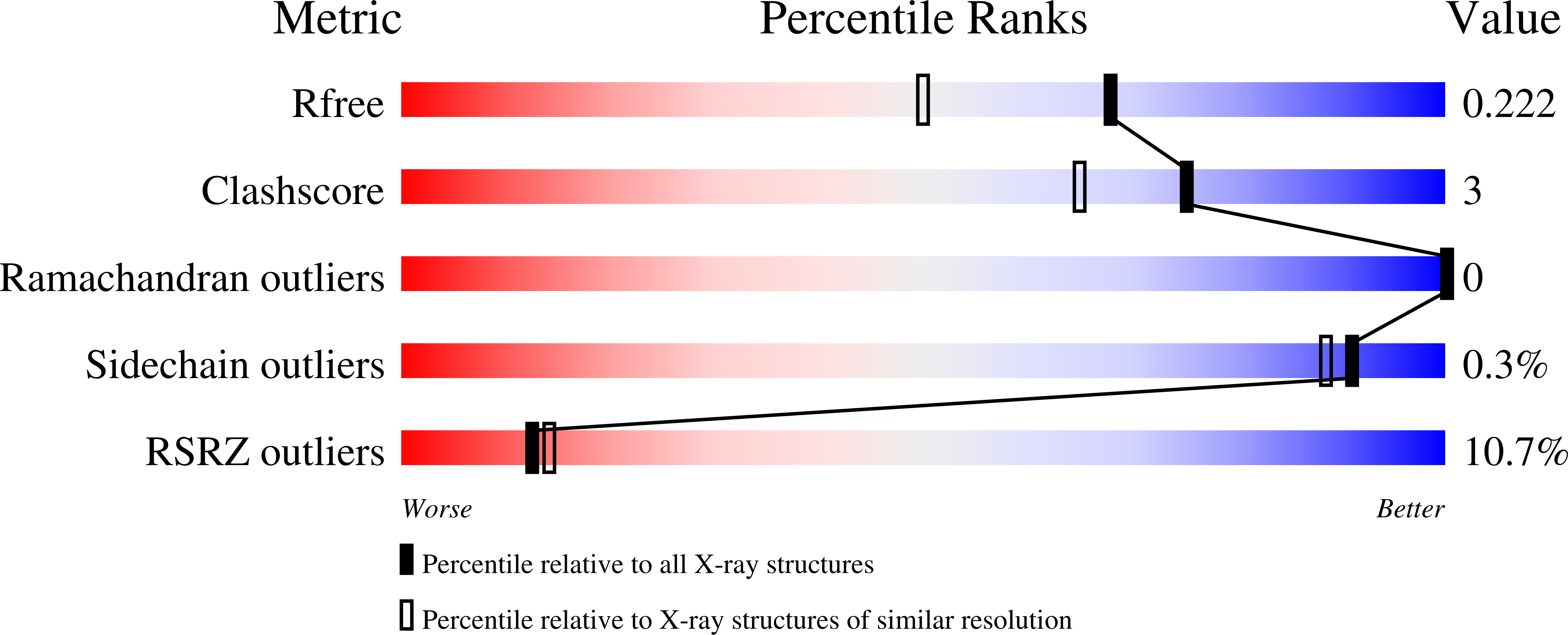
Resolution:
1.75 Å
R-Value Free:
0.22
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
C 2 2 21