Deposition Date
2024-10-08
Release Date
2025-08-06
Last Version Date
2025-11-12
Entry Detail
PDB ID:
9H0B
Keywords:
Title:
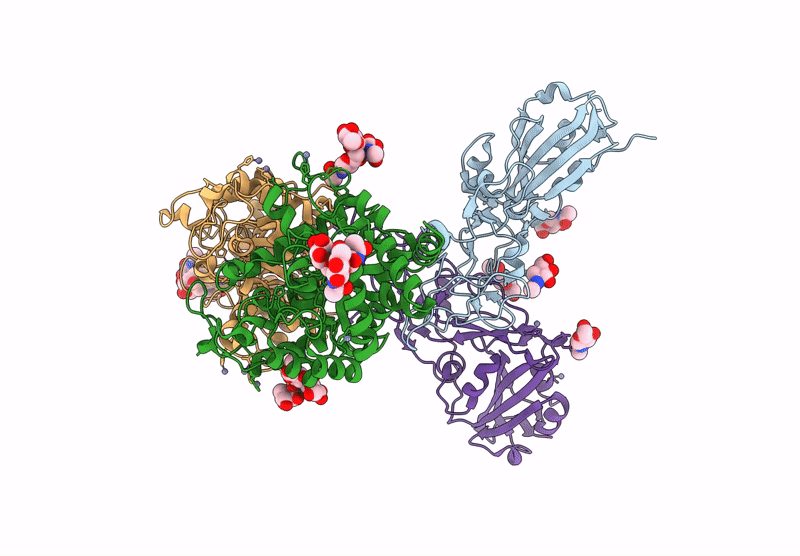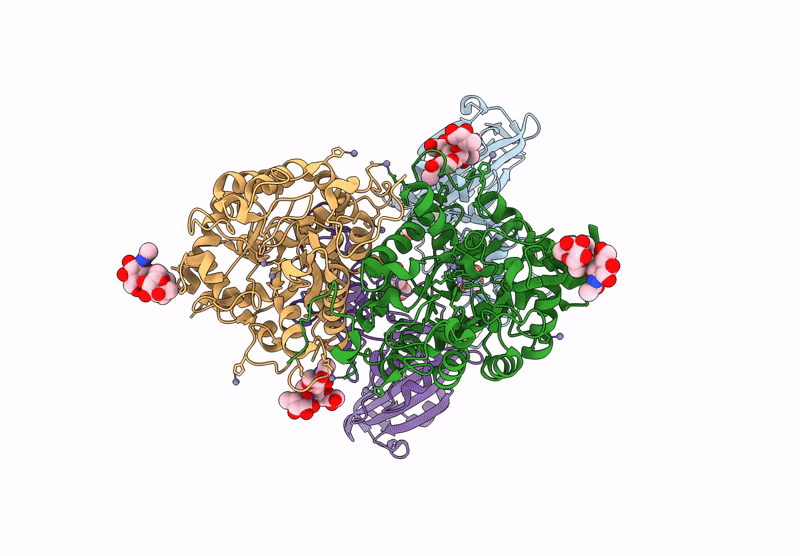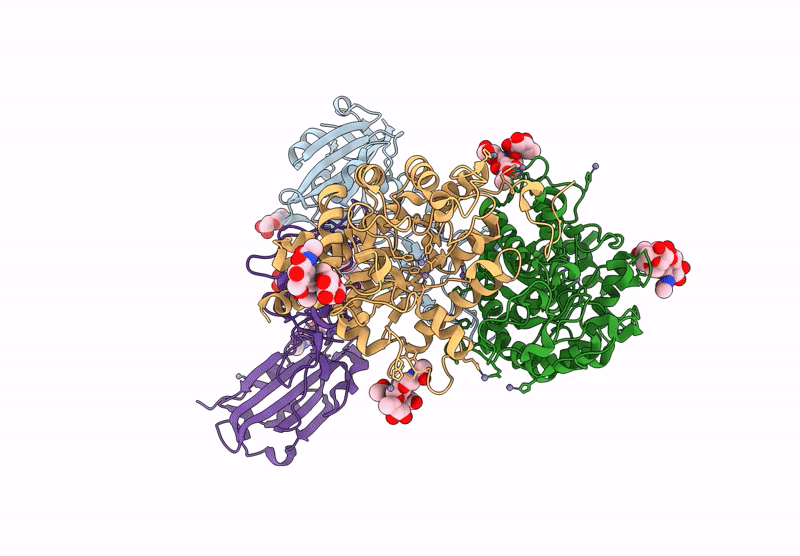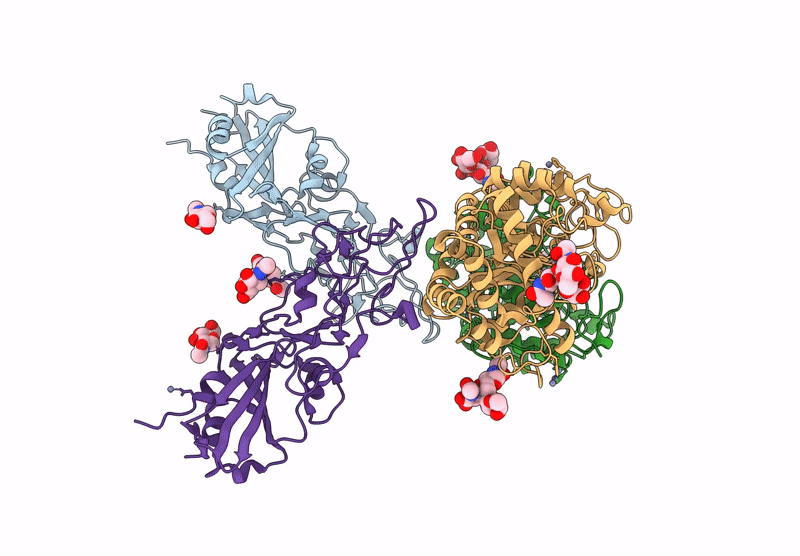
Crystal structure of the Porcine Hemagglutinating Encephalomyelitis Virus (PHEV) receptor binding domain in complex with porcine DPEP1.
Biological Source:
Source Organism:
Sus scrofa (Taxon ID: 9823)
Porcine hemagglutinating encephalomyelitis virus (Taxon ID: 42005)
Porcine hemagglutinating encephalomyelitis virus (Taxon ID: 42005)
Host Organism:
Method Details:
Experimental Method:
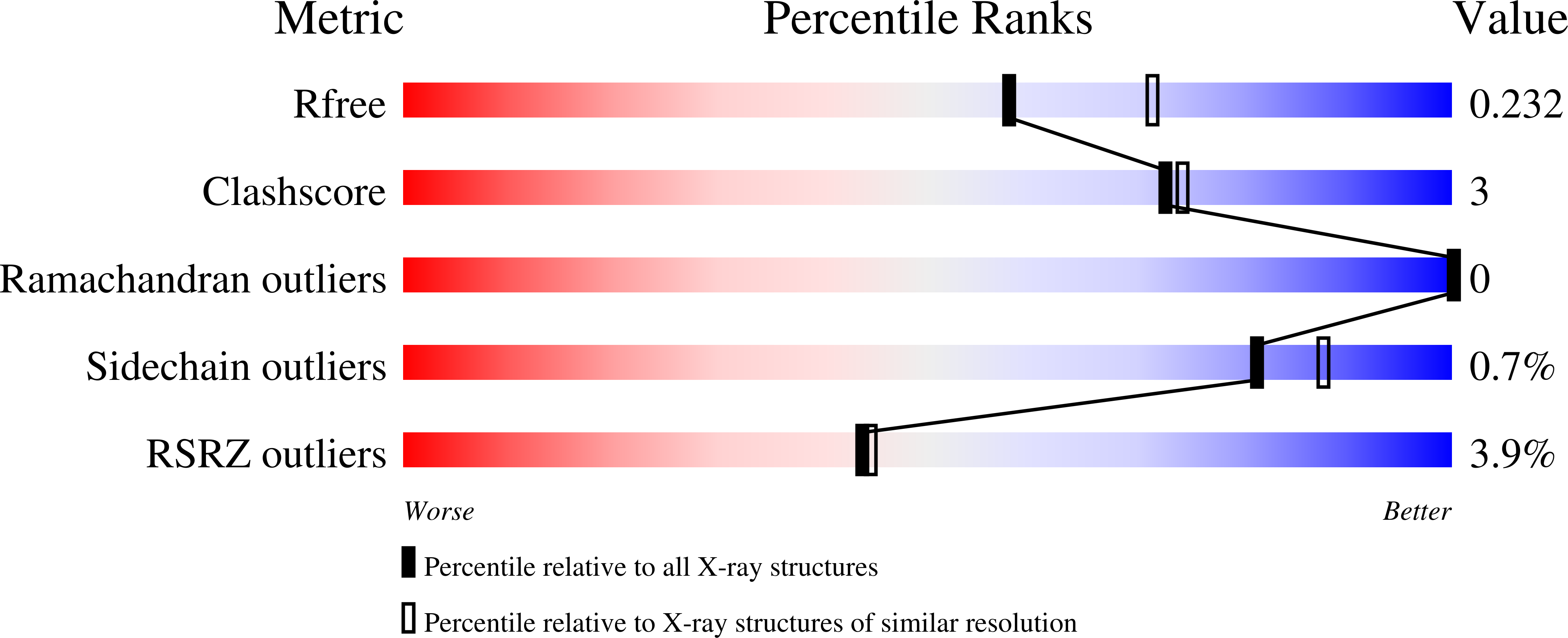
Resolution:
2.25 Å
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 1 21 1