Deposition Date
2024-09-09
Release Date
2025-01-15
Last Version Date
2025-03-19
Entry Detail
PDB ID:
9GQK
Keywords:
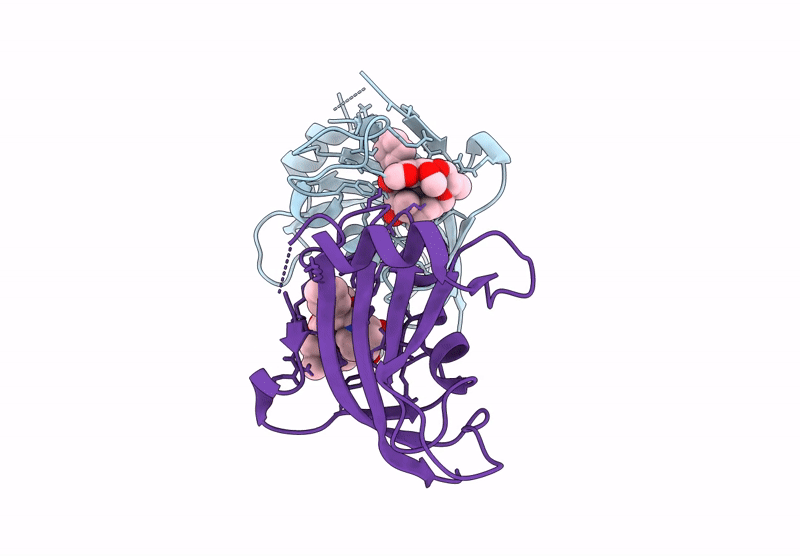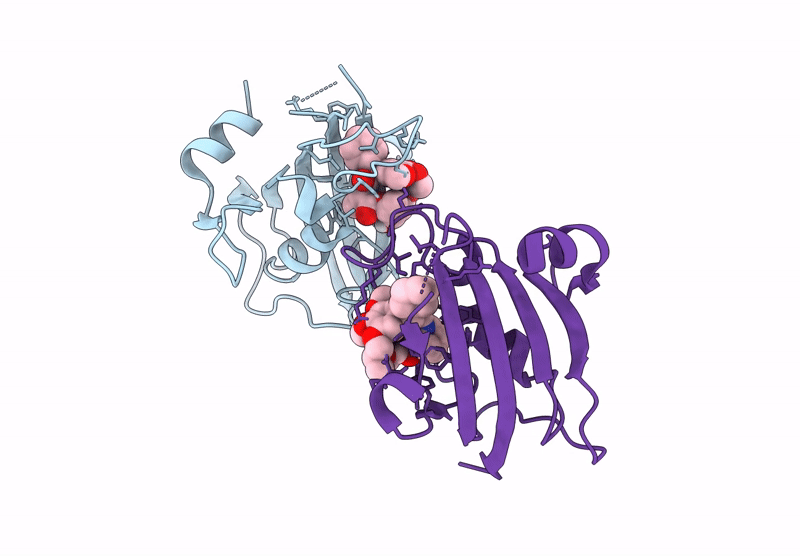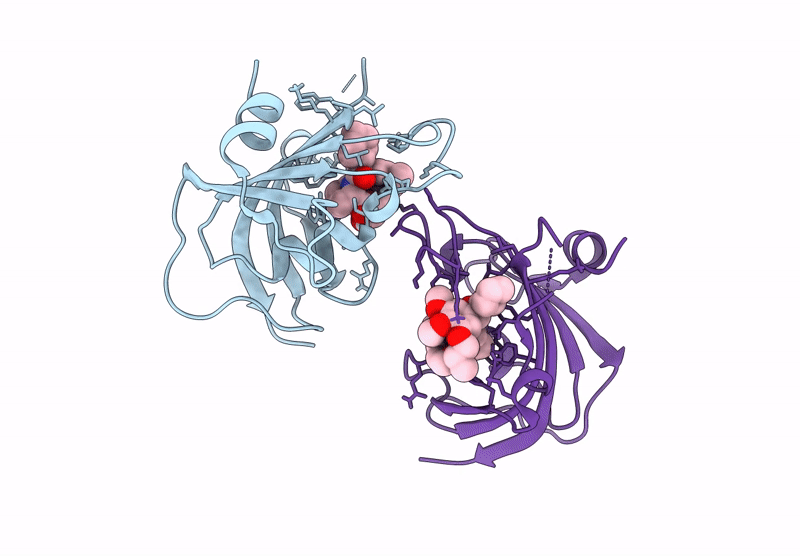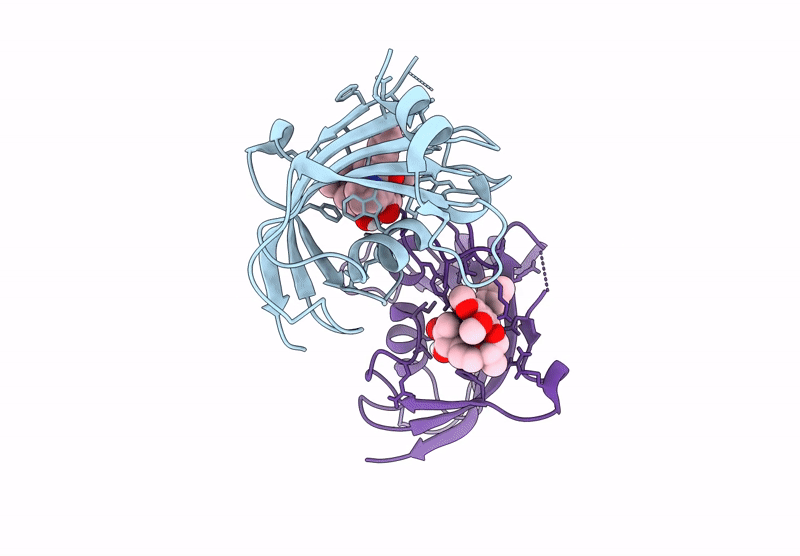
Title:
The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog m5(11,5)-(E)-OH
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.70 Å
R-Value Free:
0.28
R-Value Work:
0.23
Space Group:
P 32 2 1


