Deposition Date
2024-07-17
Release Date
2025-06-18
Last Version Date
2025-06-18
Entry Detail
PDB ID:
9G5M
Keywords:
Title:
N-Acyl-D-amino-acid deacylase (D-acylase) from Klebsiella pneumoniae in an open conformation
Biological Source:
Source Organism(s):
Klebsiella pneumoniae subsp. pneumoniae Kp13 (Taxon ID: 1123862)
Expression System(s):
Method Details:
Experimental Method:
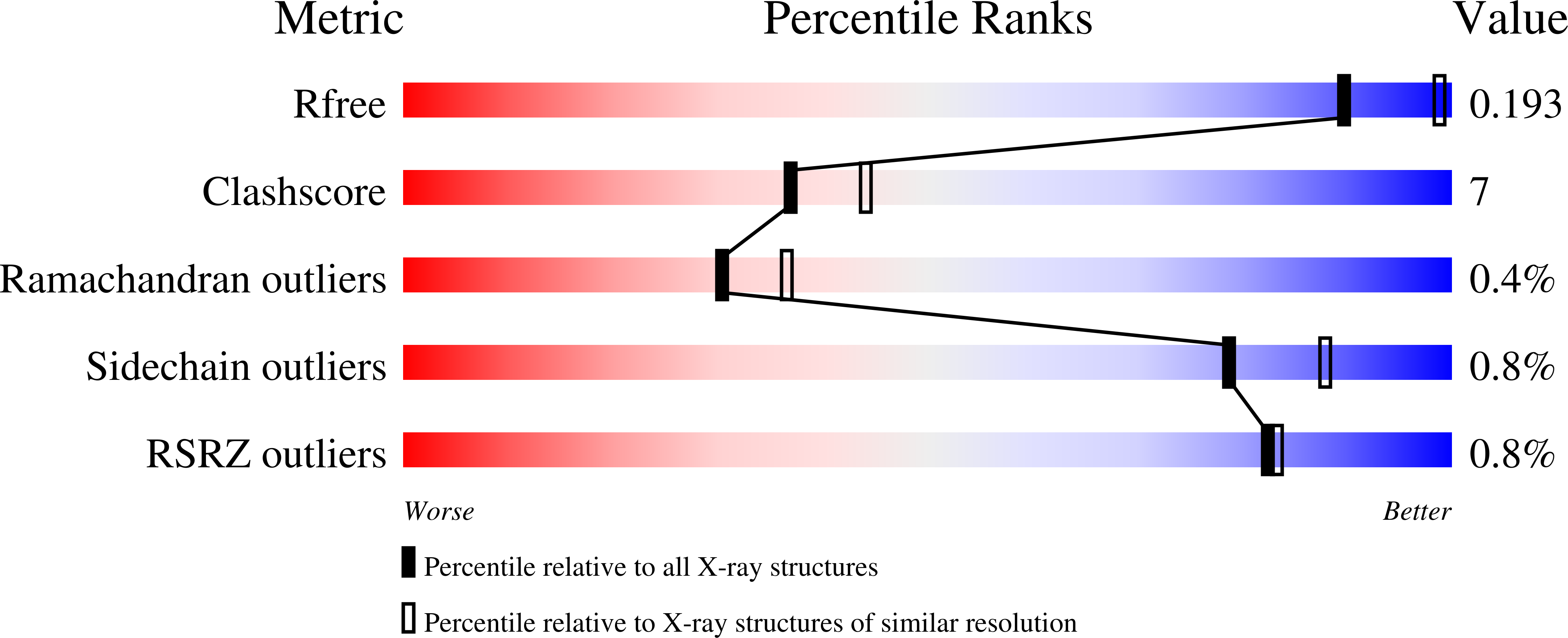
Resolution:
2.27 Å
R-Value Free:
0.19
R-Value Work:
0.15
Space Group:
P 32 2 1