Deposition Date
2024-07-15
Release Date
2025-11-05
Last Version Date
2025-12-10
Entry Detail
PDB ID:
9G4I
Keywords:
Title:
Group II intron assembly intermediate Domain 1 and 2 "Partly open" state
Biological Source:
Source Organism(s):
Oceanobacillus iheyensis (Taxon ID: 182710)
Method Details:
Experimental Method:
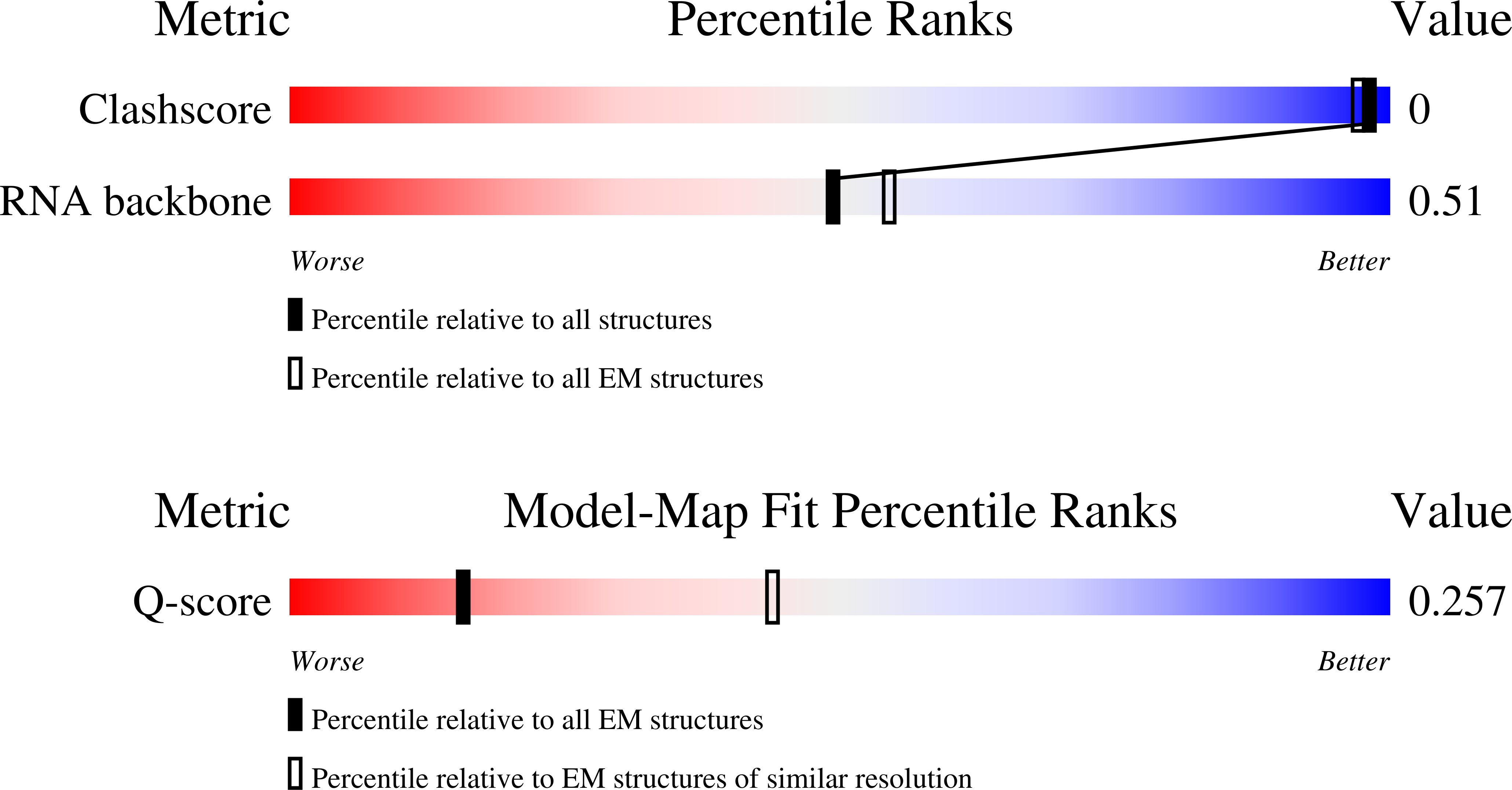
Resolution:
4.61 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE