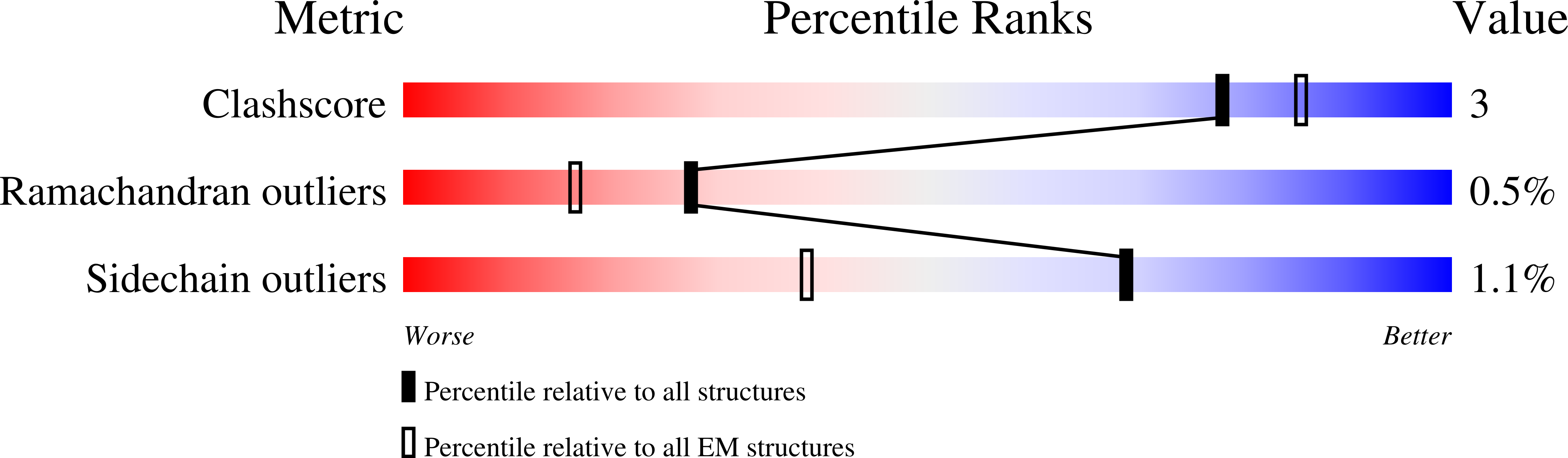
Deposition Date
2024-07-07
Release Date
2025-07-16
Last Version Date
2025-07-16
Entry Detail
PDB ID:
9G05
Keywords:
Title:
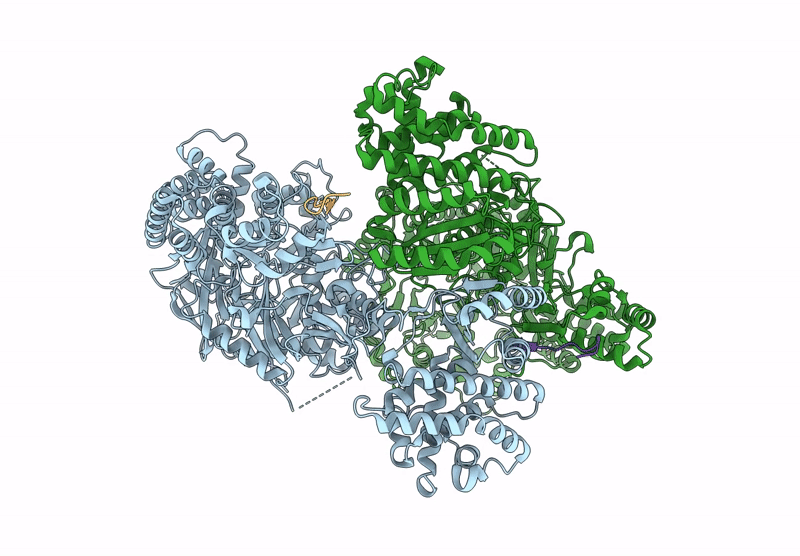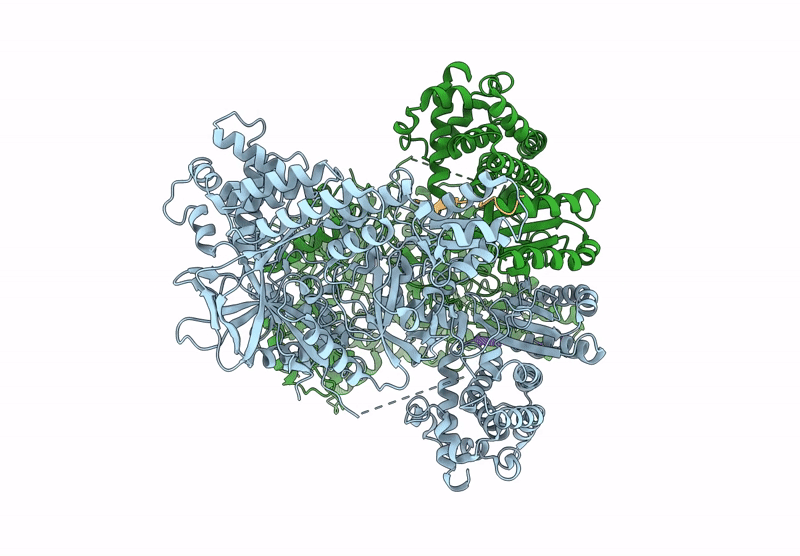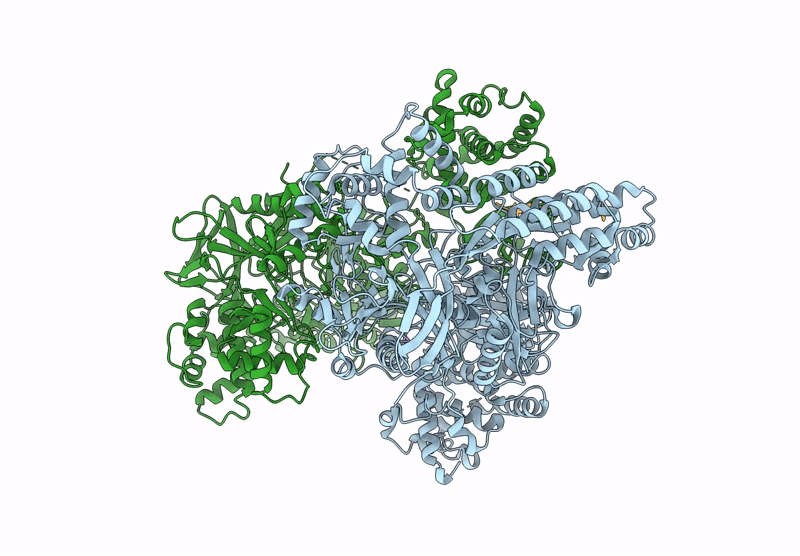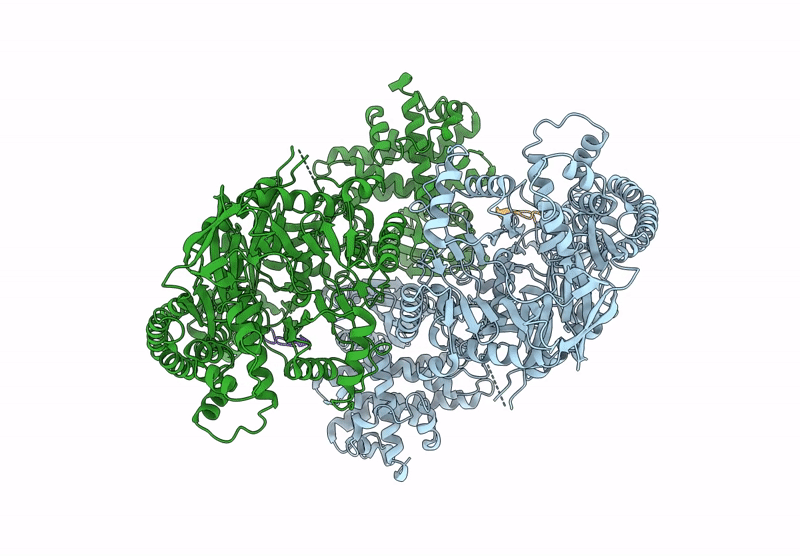
Structure of MadB, a class I dehydrates from Clostridium maddingley, in complex with its substrate
Biological Source:
Source Organism(s):
Clostridium sp. Maddingley MBC34-26 (Taxon ID: 1196322)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.13 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE