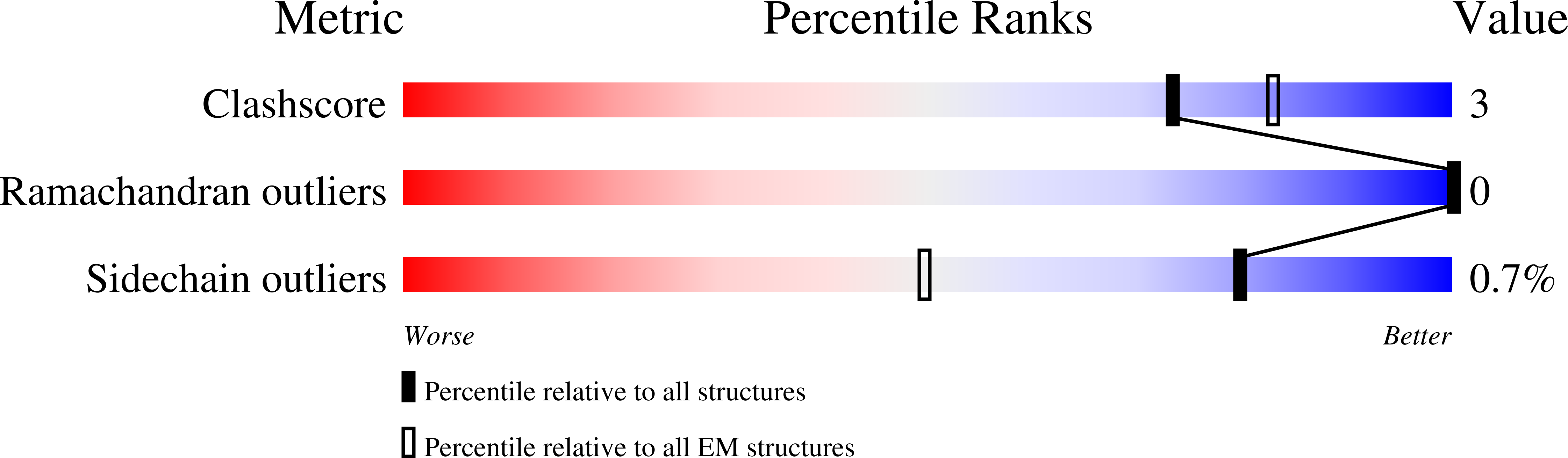
Deposition Date
2024-04-09
Release Date
2025-04-23
Last Version Date
2025-11-05
Entry Detail
PDB ID:
9EYM
Keywords:
Title:
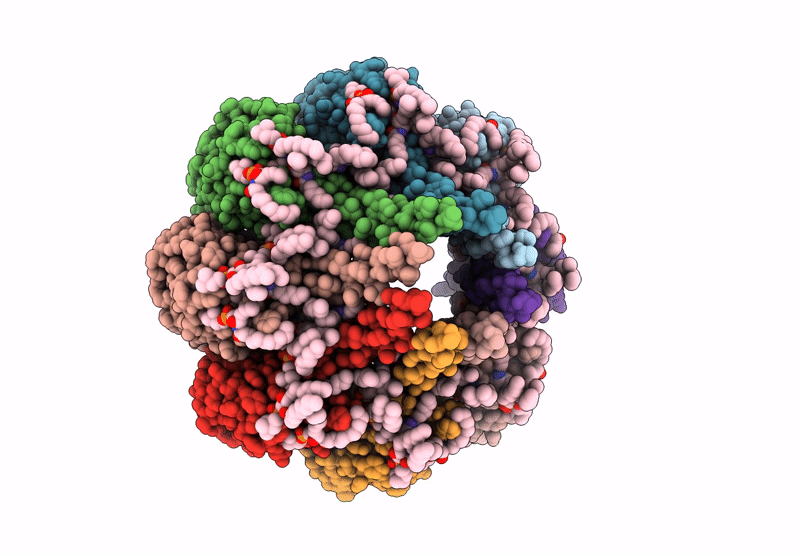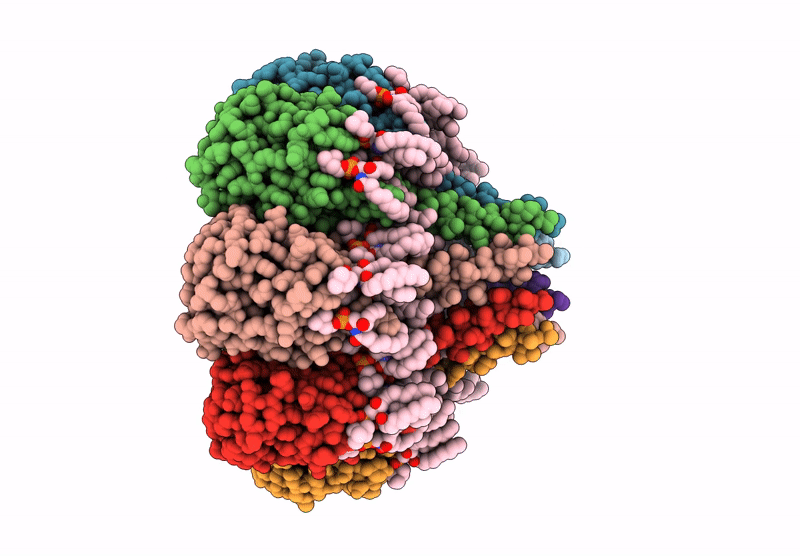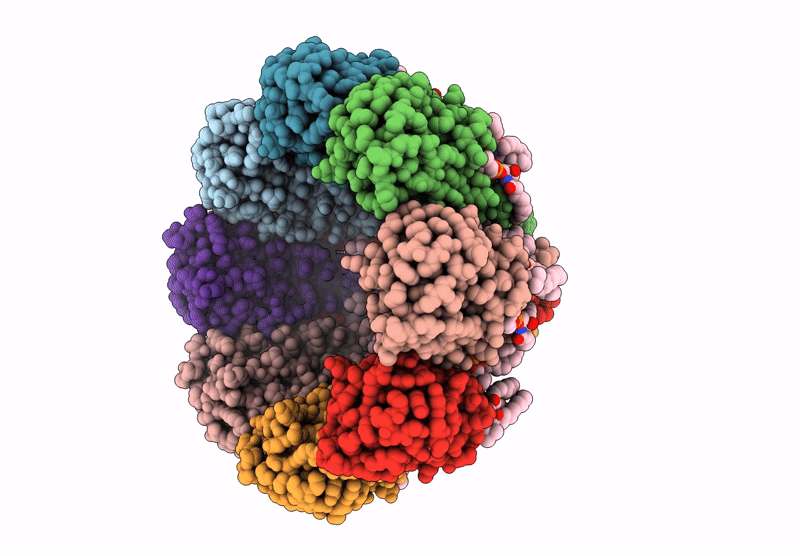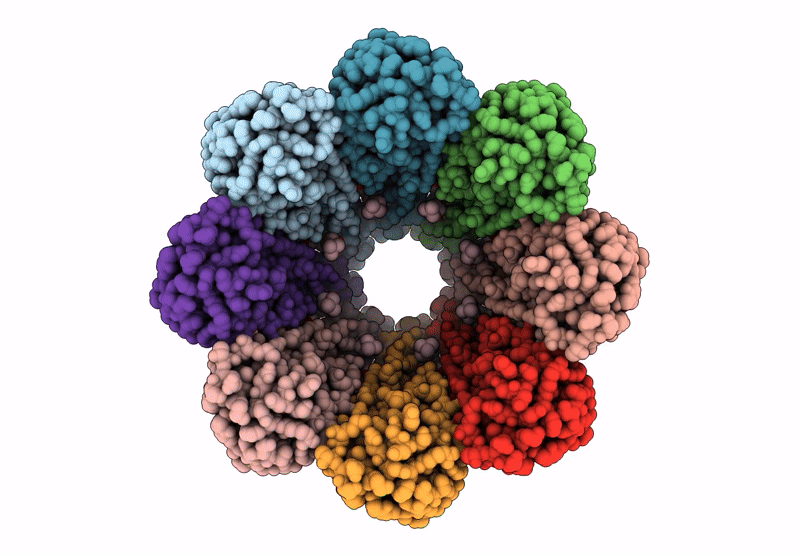
The structure of solubilized octameric pore of actinoporin Fav prepared on DOPC:sphingomyelin membranes
Biological Source:
Source Organism(s):
Orbicella faveolata (Taxon ID: 48498)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.60 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE