Deposition Date
2024-03-29
Release Date
2025-08-06
Last Version Date
2025-09-17
Entry Detail
PDB ID:
9EVG
Keywords:
Title:
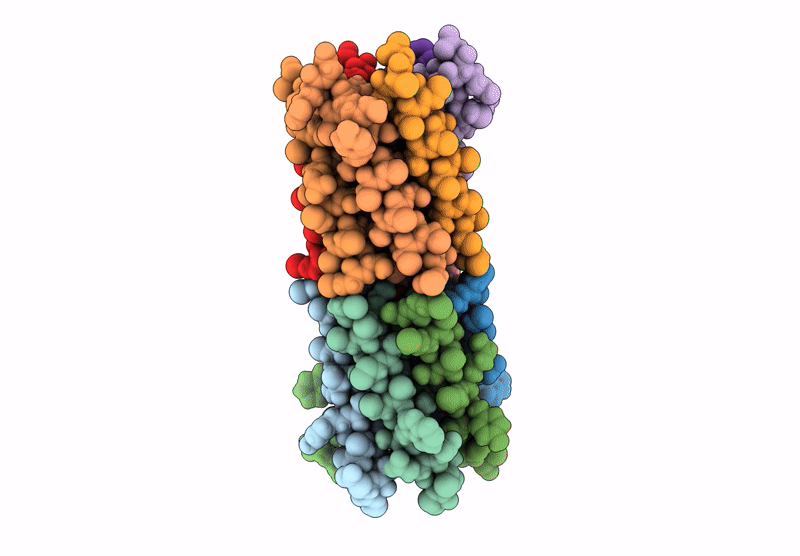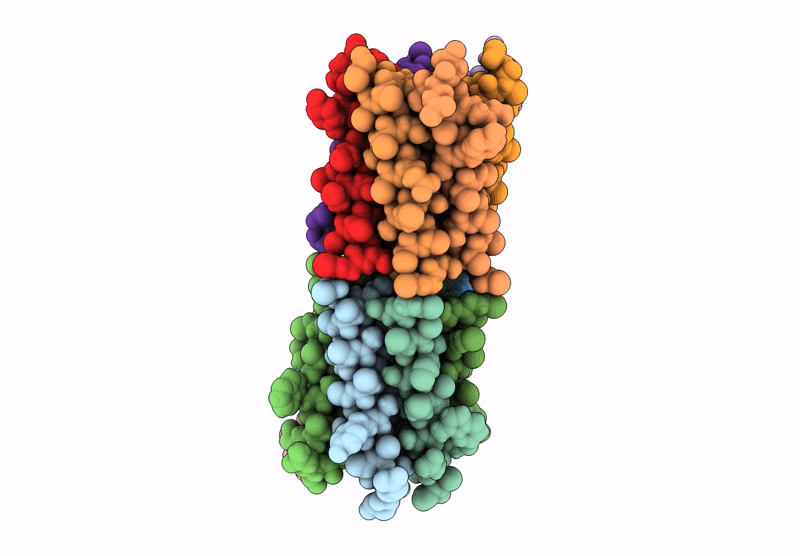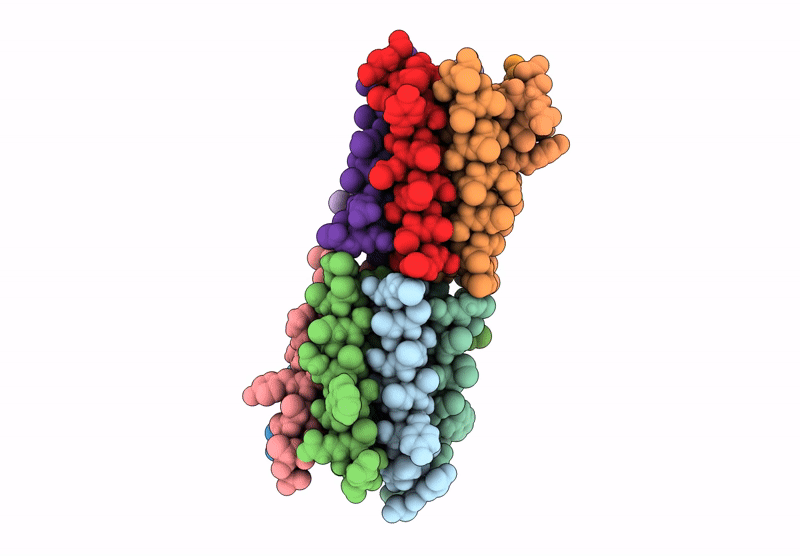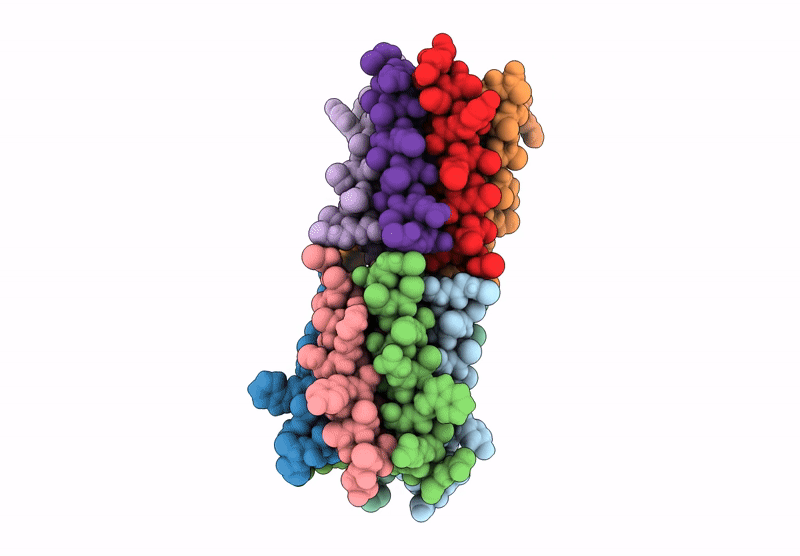
X-ray crystal structure of a de novo designed parallel coiled-coil heterohexamer with 3 heptad repeats, CCHex2-AB-g
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
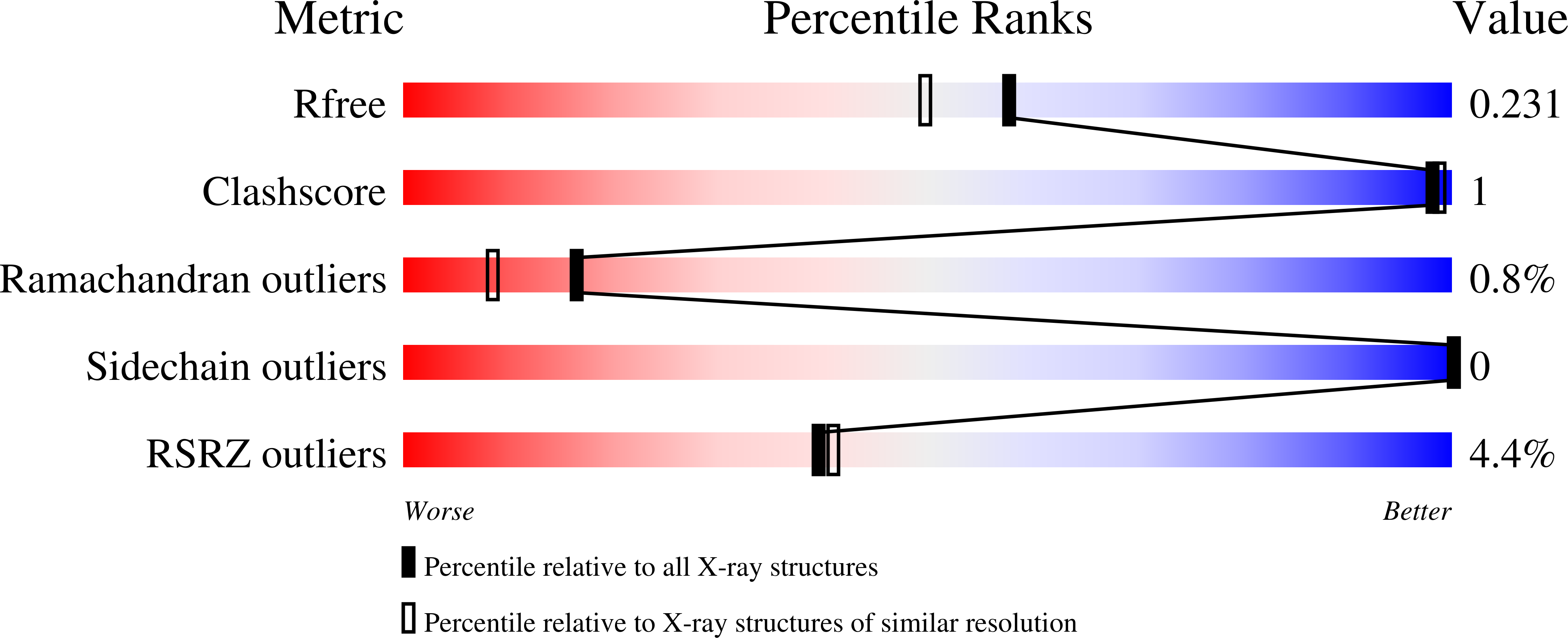
Resolution:
1.90 Å
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 1 21 1