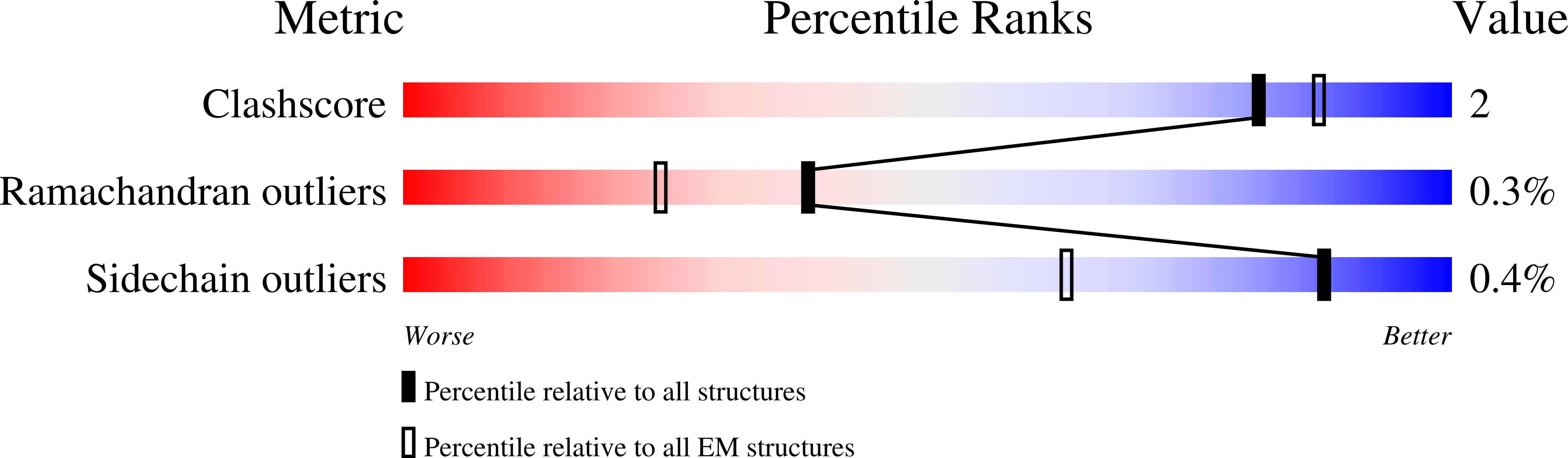
Deposition Date
2024-03-29
Release Date
2024-09-18
Last Version Date
2024-09-18
Entry Detail
PDB ID:
9EVD
Keywords:
Title:
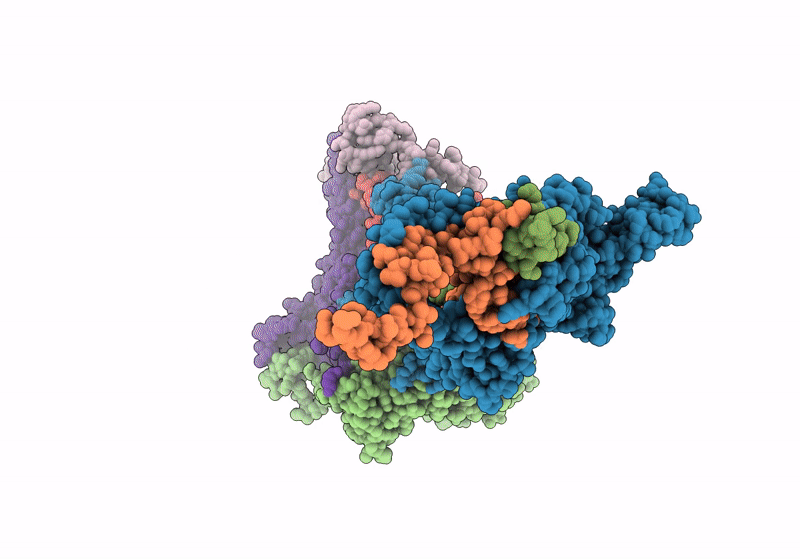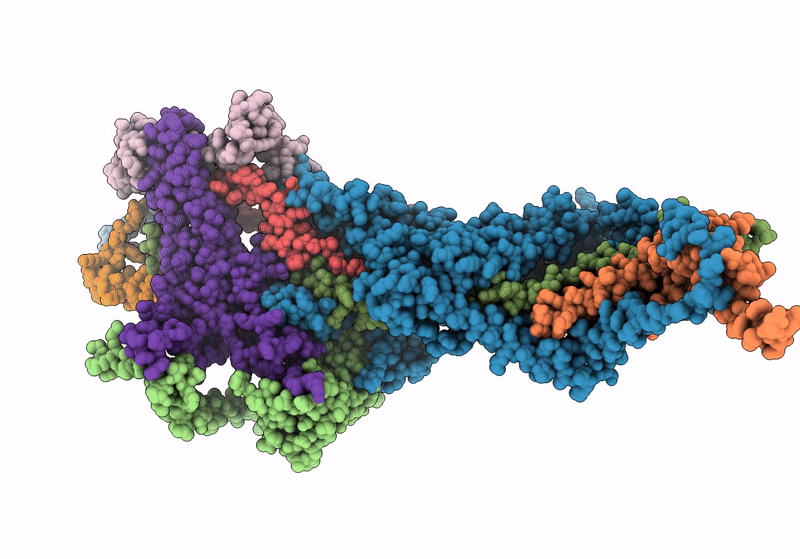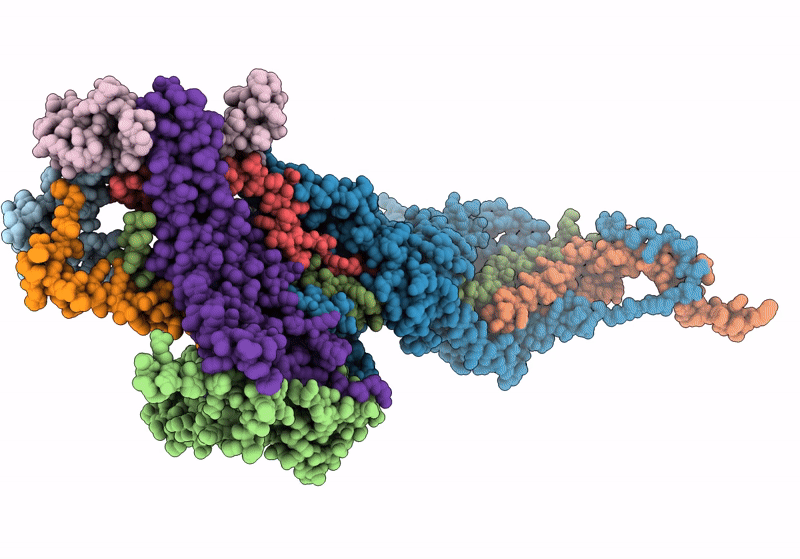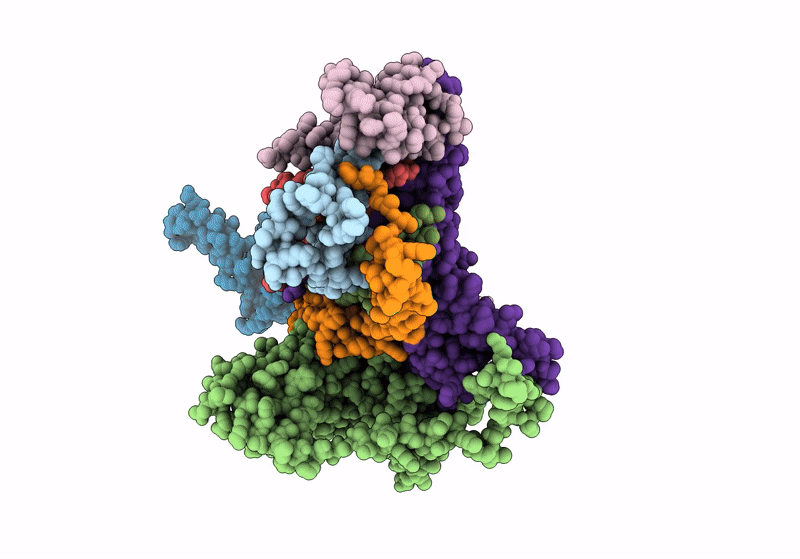
In situ structure of the peripheral stalk of the mitochondrial ATPsynthase in whole Polytomella cells
Biological Source:
Source Organism(s):
Polytomella sp. Pringsheim 198.80 (Taxon ID: 37502)
Method Details:
Experimental Method:
Resolution:
5.60 Å
Aggregation State:
CELL
Reconstruction Method:
SUBTOMOGRAM AVERAGING