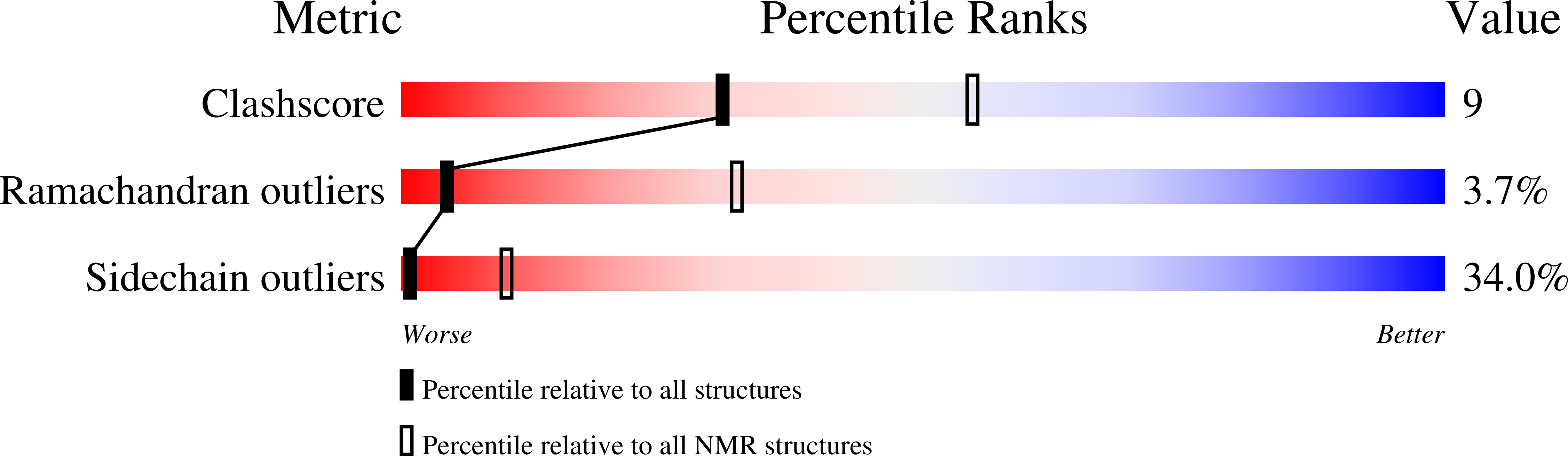
Deposition Date
2024-11-12
Release Date
2025-05-07
Last Version Date
2025-07-09
Entry Detail
PDB ID:
9EBE
Keywords:
Title:
Solution structure of sigma-S-GVIIIA conotoxin extracted from Conus geographus
Biological Source:
Source Organism(s):
Conus geographus (Taxon ID: 6491)
Method Details:
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
20
Selection Criteria:
target function