Deposition Date
2024-11-05
Release Date
2024-12-04
Last Version Date
2025-12-24
Entry Detail
PDB ID:
9E8I
Keywords:
Title:
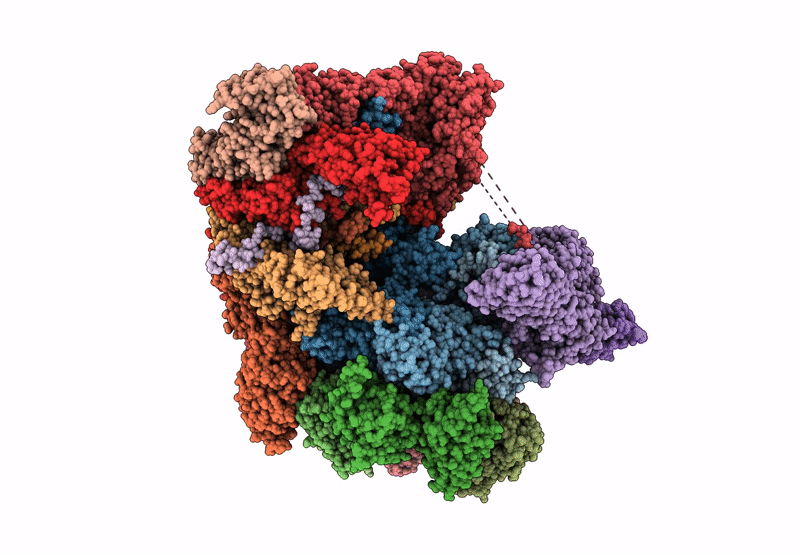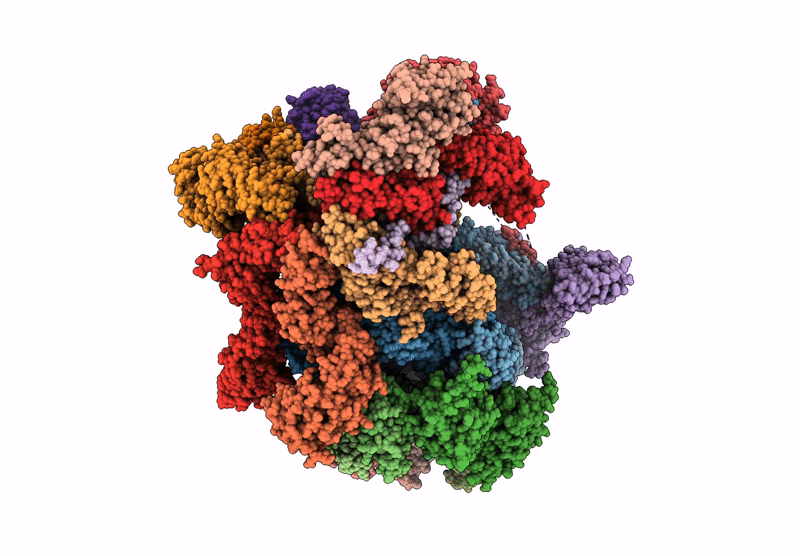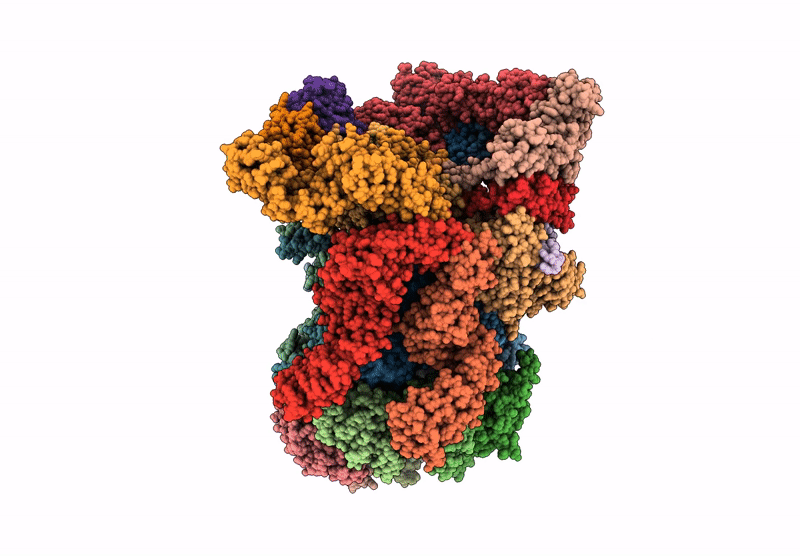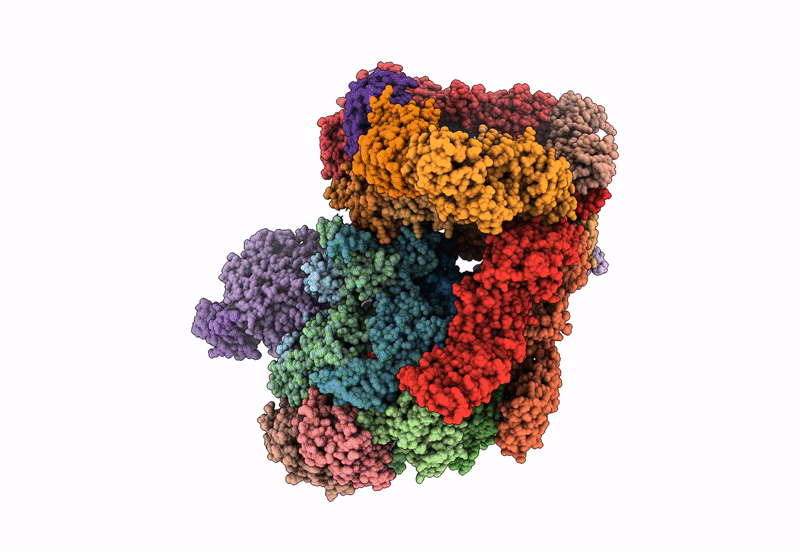
Human proteasome in resting state conformation bound to TXNL1 in Forward conformation
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.87 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


